RNAscope common questions, troubleshooting, and tips and tricks
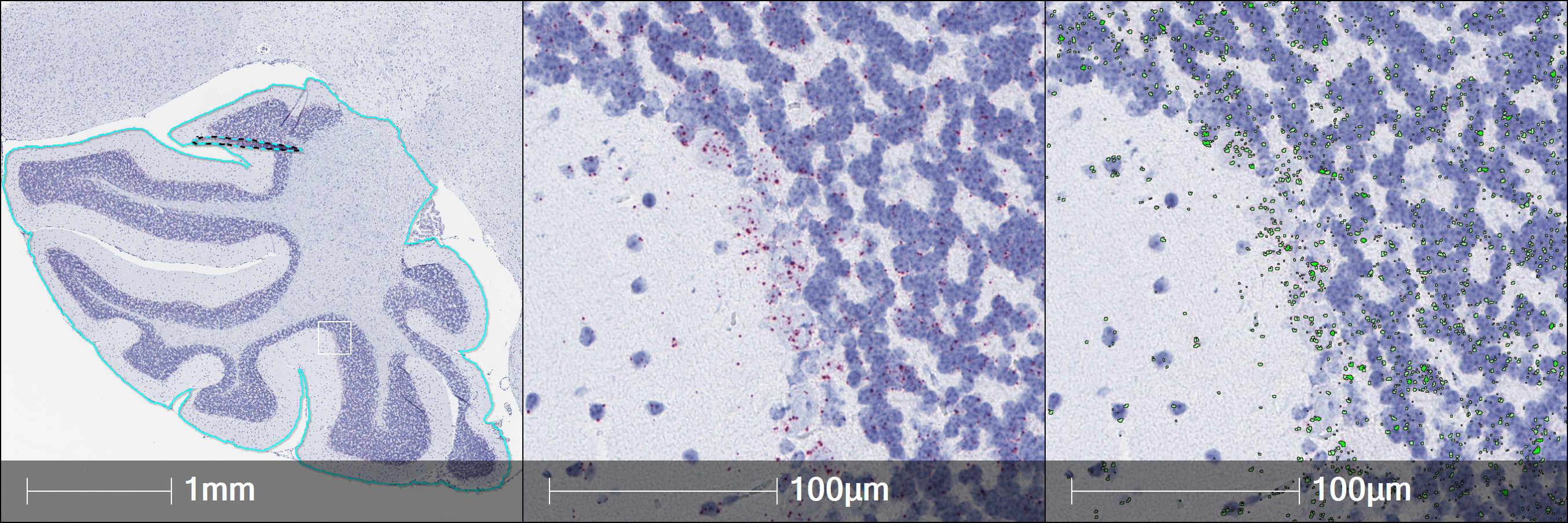
When performing RNAscope image analysis, you can set yourself up for success with a combination of thoughtful experimental design, rigorous assay execution and validation. At Indica Labs, the most common challenges we see in RNAscope image analysis include chromogenic staining that has saturated to black, which causes color deconvolution challenges, loss of nuclear morphology due to over-digestion of tissue, and failing to verify and validate an image analysis solution before scaling up.
Check out our RNAscope FAQ listed below and download our recent Quantitative RNAscope Image Analysis Guide to learn more about each of the most common challenges and how to avoid them.
Q: Should I capture my RNAscope images at 20x or 40x magnification?
A: Image acquisition for RNAscope images is recommended at 40x magnification.
Q: I’m not sure how to optimize the parameters for my RNAscope module in HALO®. Where do I start?
A: Sign up for the Indica Labs Learning Portal, navigate to your module of interest, download the User Guide, and watch the training tutorial video. Watch the RNAscope Masterclass webinar and reach out to support@indicalab.com for a refresher training on your modules.
Q: How can I eliminate tissue artifacts from my image analysis?
A: Manual annotation tools are available to eliminate one off artifacts in image analysis. Use the Exclusion tool (scissors) to draw an exclusion layer, or hold ctrl down while using the Magnetic pen, Brush tool, etc. These tools are also handy to remove areas where tissue has folded back on itself. If you need to remove the tissue edge artifacts, use the Tissue Edge Thickness parameter in the Advanced Analysis menu. Finally, the Tissue Classifier or one of the HALO AI™ neural network tissue classification algorithms provide additional ways to remove artifacts, such as tissue folds.
Q: How do you manage artifacts that negatively impact spot counting?
A: Any time a distinct color can be excluded without impacting stains of interest, the Exclusion Stain is recommended. If you have two problematic features, such as anthracotic pigments (carbon deposits) in lung tissue and red blood cells, you can use HALO AI or the HALO Tissue Classifier module to detect and exclude the red blood cells and Exclusion Stain to remove the anthracotic pigments.
Q: How do I manage heterogeneous ISH staining patterns, such as PD-L1?
A: If you have morphologically distinct regions, it may be helpful to use the HALO AI and/or Tissue Classifier to isolate tissues of interest for analysis. Annotations can also be drawn manually.
Q: What do I do if it is difficult to distinguish my RNAscope signal from background?
A: See the following documentation for staining optimization: RNAscope TroubleShooting Guide.
Q: What do I do if my experimental sample has no signal?
A: Confirm that both your positive and negative controls score as expected before making any conclusions on your experimental sample. Confirm that you are using the POLR2A positive control probe for low expression assays.
If you have additional questions, reach out to support@indicalab.com to reach our global Applications and Technical Solutions teams at Indica Labs for image analysis support. Reach out to info@indicalab.com to learn more about HALO and HALO AI image analysis solutions and to reach our Pharma Services team to explore end-to-end pathology research project support.