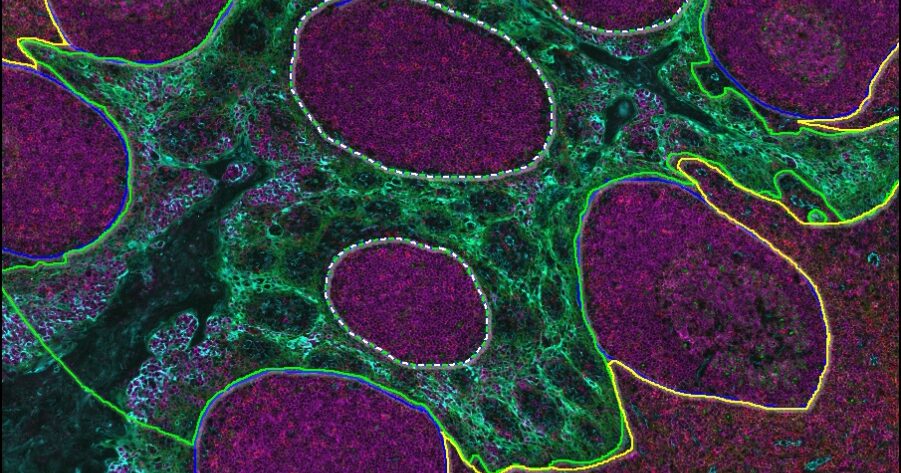
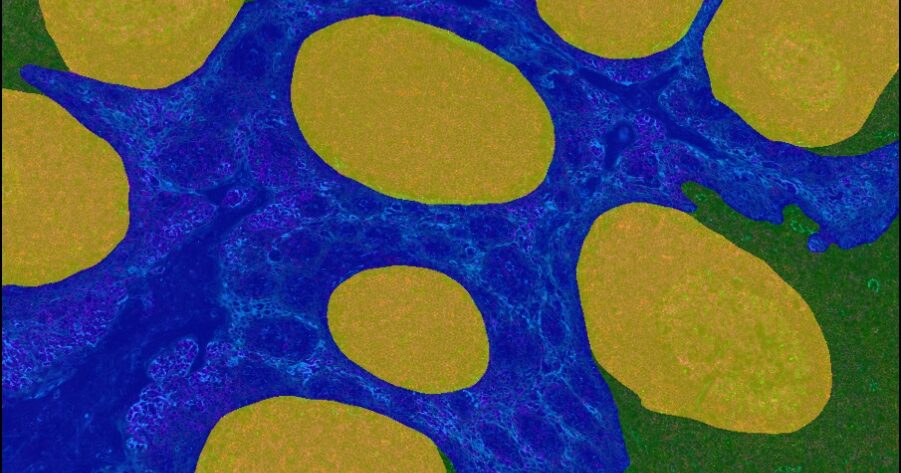
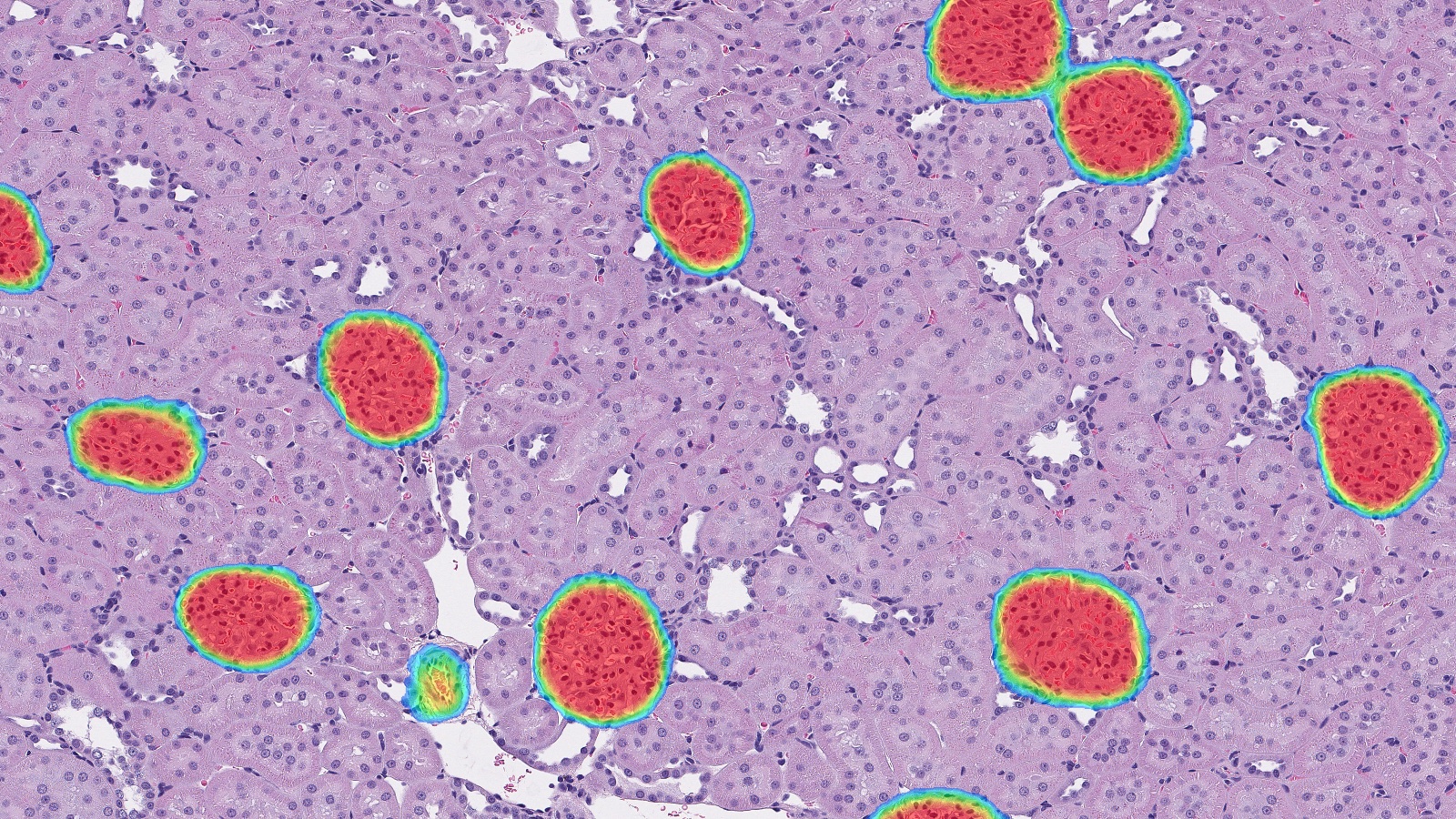
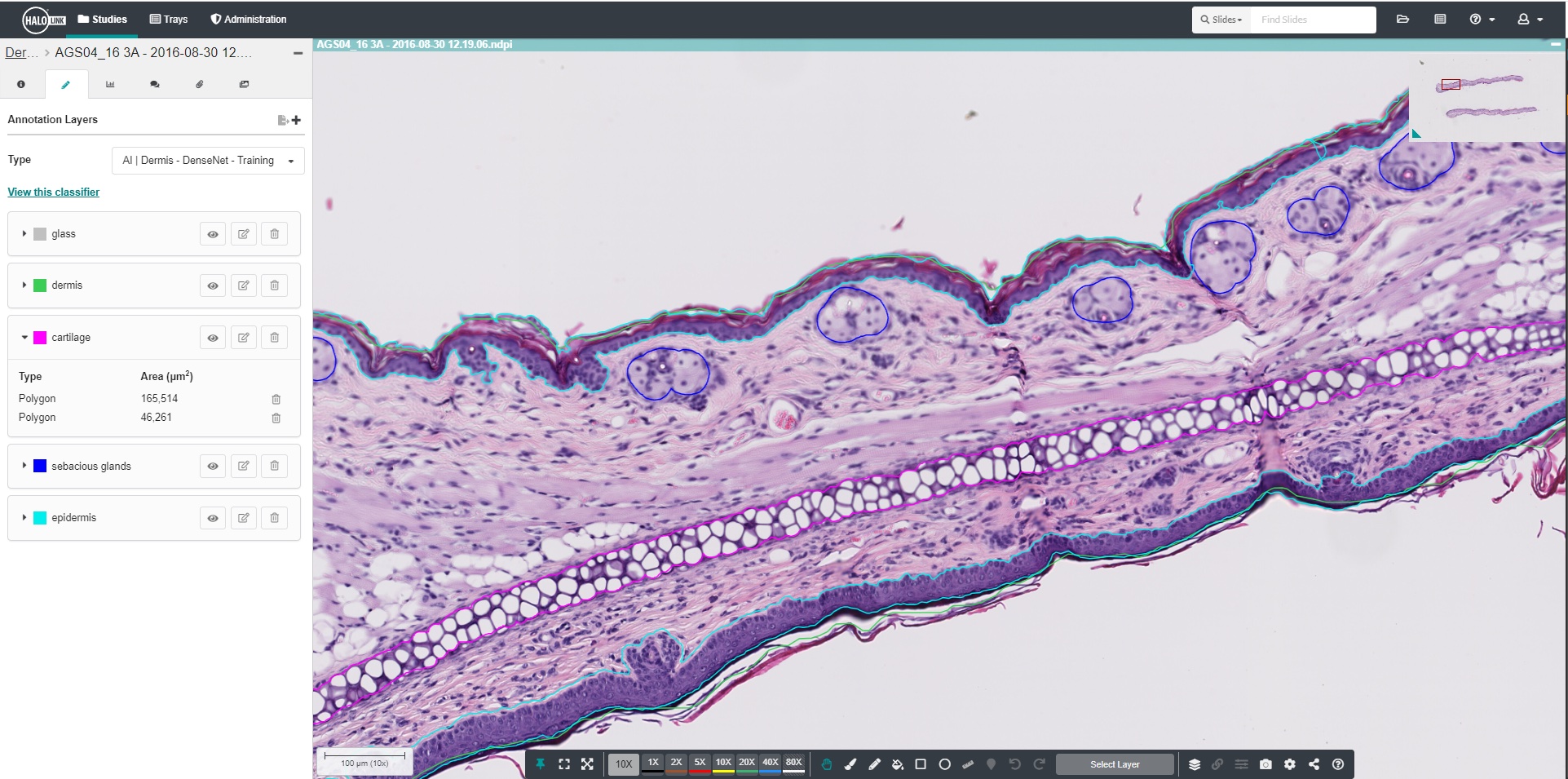
Advancing Discovery with Trainable AI
Insightful and robust AI creation for pathology


Unlock Limitless Possibilities with HALO AI
HALO AI has you covered. Whether you’re looking to solve image analysis challenges in segmentation, classification, or phenotyping, the advanced deep learning networks of HALO AI are easily tuned for your specific application with the train-by-example interface.
Diverse and Powerful AI
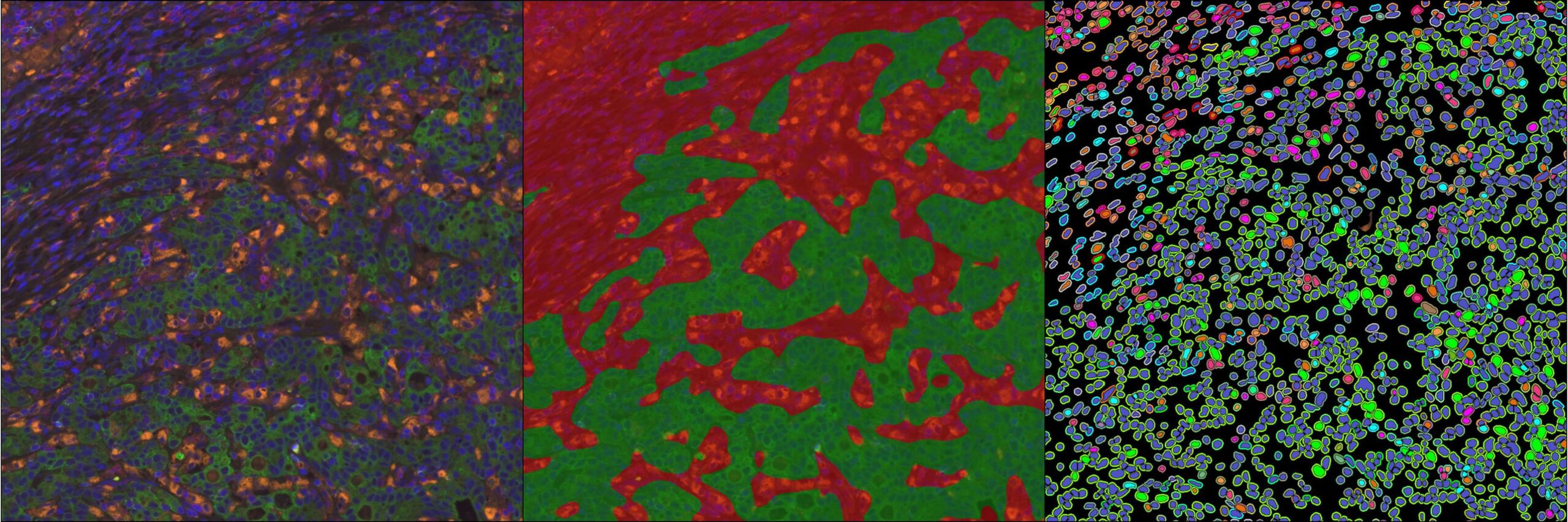
Underpinned by modern deep learning networks, HALO AI provides a collection of segmentation, classification, and phenotyping tools for brightfield and fluorescence applications. Train a classifier to quantify tissue classes, to find rare events or cells in tissues, or to categorize cell populations into specific phenotypes.
AI-Powered Annotation
Use a point and click workflow to rapidly annotate images and develop training data.
Nuclear Segmentation

Choose between several trainable AI-based nuclear segmentation networks to optimize segmentation accuracy when nuclear morphologies vary.
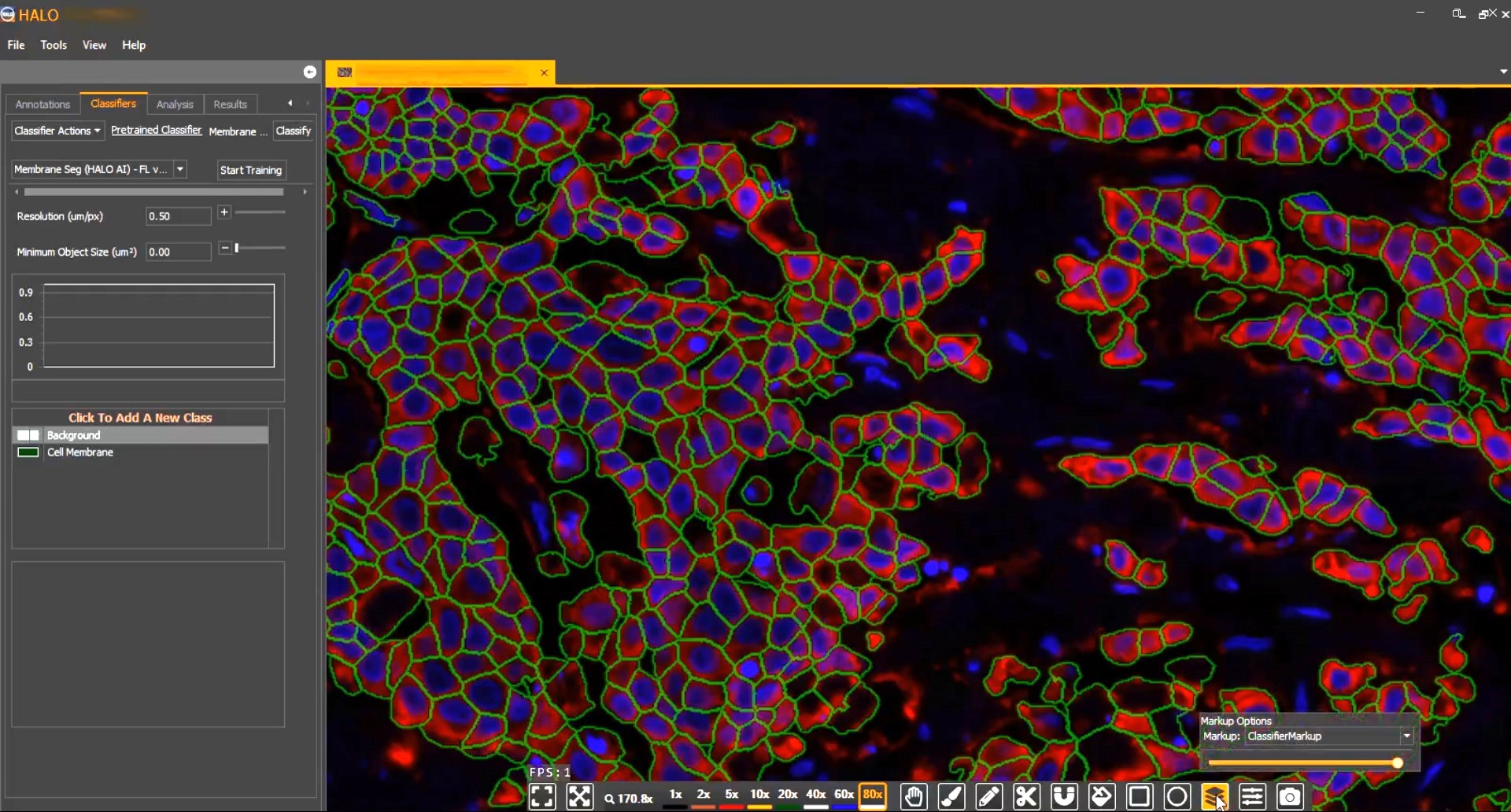
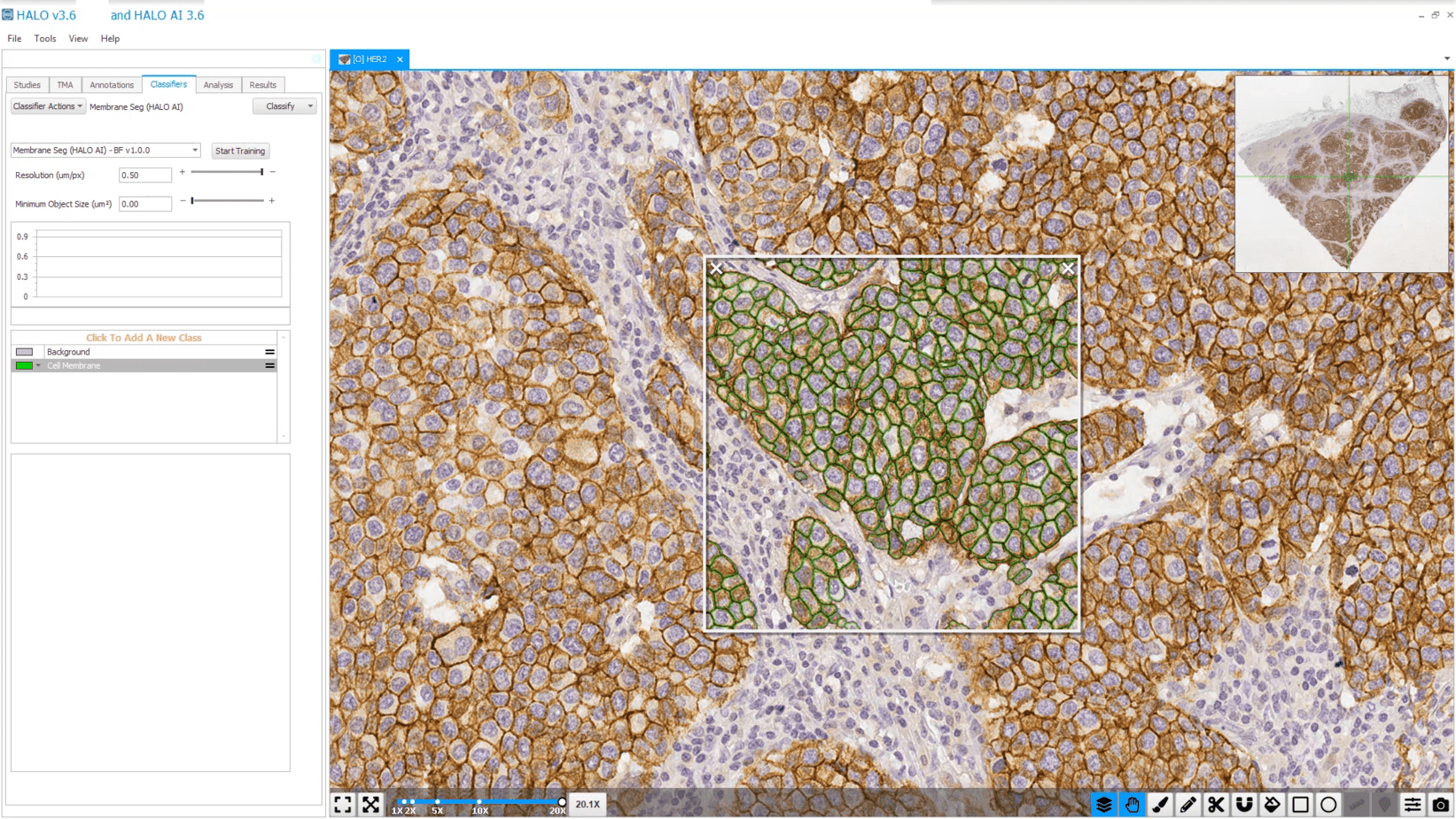
Membrane Segmentation

Train AI-powered membrane segmentation networks to accurately delineate cell membranes for your specific assay.
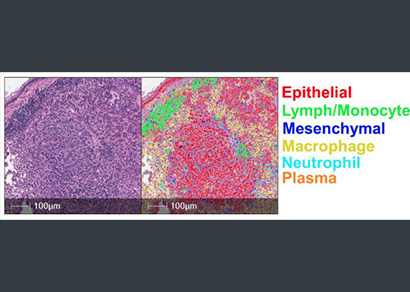
Cell & Object Phenotyping

Quickly train a phenotyper to quantify cell types or objects of interest.
Slide Quality Control

Leverage a trainable AI-powered quality control network to detect common artifacts in H&E and IHC images.
Simple & Intuitive Workflow
Choose to integrate your trained HALO AI networks in multiple locations within HALO modules to perform functions including tissue classification, nuclear and membrane segmentation, and phenotyping. Alternatively, apply HALO AI directly to an entire study of whole slide images or to select regions of interest – the choice is yours.
Manage Variability
Extreme variability runs rife in image analysis but is no match for HALO AI. Common sources of variability include diverse morphologies, alterations of morphologies from staining protocols, differences in tissue quality, uneven staining, and more.
HALO AI can be easily trained to accommodate variability to deliver accurate segmentation and classification results across large studies. HALO AI can even be trained to work across vastly different stains such as PAMS, Trichrome, H&E, and IHC.
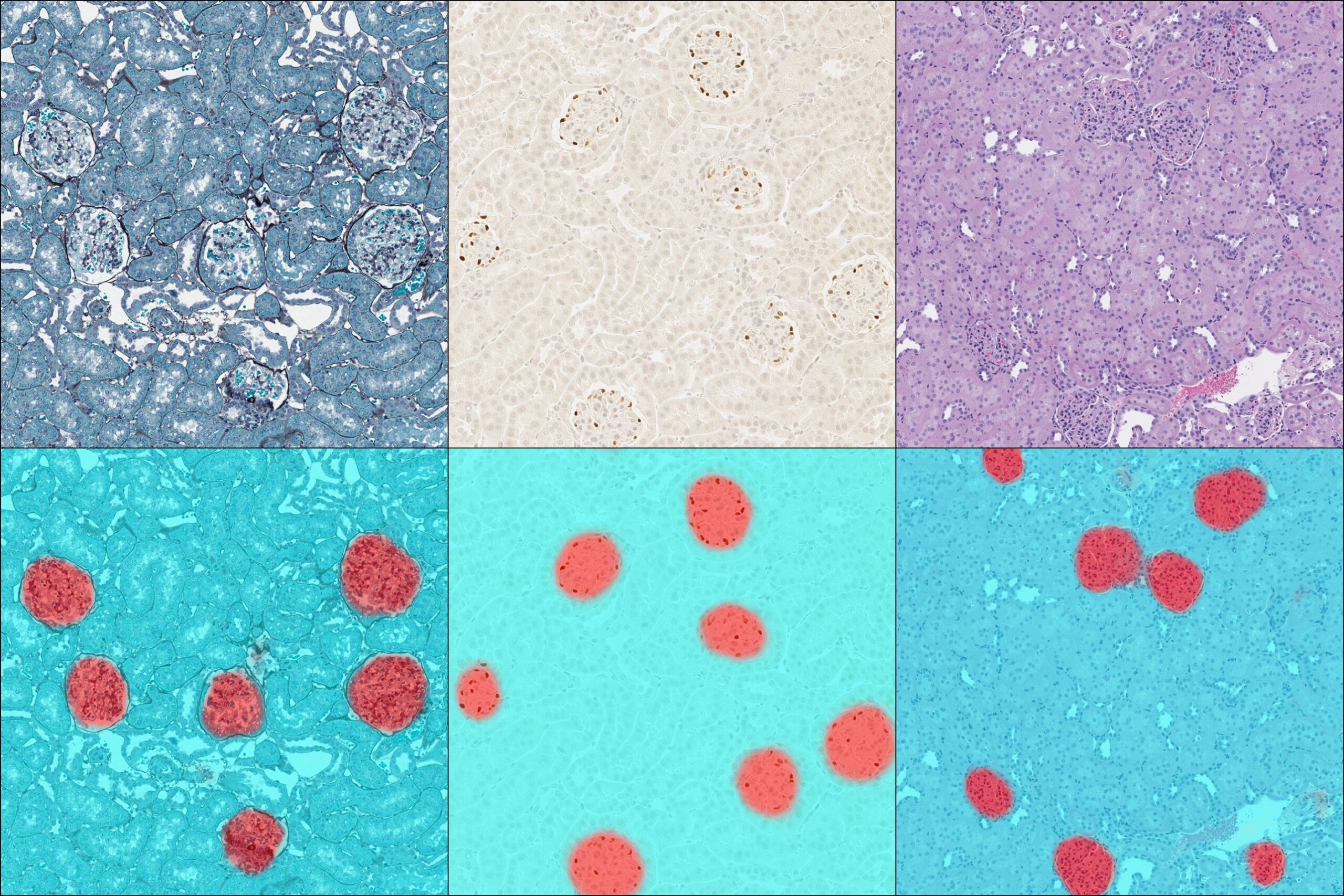
Exceptional Identification
Pre-trained nuclear and membrane segmentors available in HALO are advanced tools for nuclear and membrane segmentation, but when you need to optimize a network for a bespoke application, you need HALO AI.
As with the tissue classification and segmentation networks, you can quickly add training data with the AI-based annotation tool and train the network to optimally segment nuclei or cell membranes. And once trained, any HALO AI network can be incorporated into HALO modules for maximum utility.

Powerful Tissue Classification
The degree of variability encountered in pathology often demands an approach above and beyond a simple random forest tissue classifier.
HALO AI has two advanced pre-trained networks capable of creating high-resolution classifiers in brightfield and fluorescence, depending on how much training data and time are available for training and optimization.
Curious to see HALO AI in action on your images?
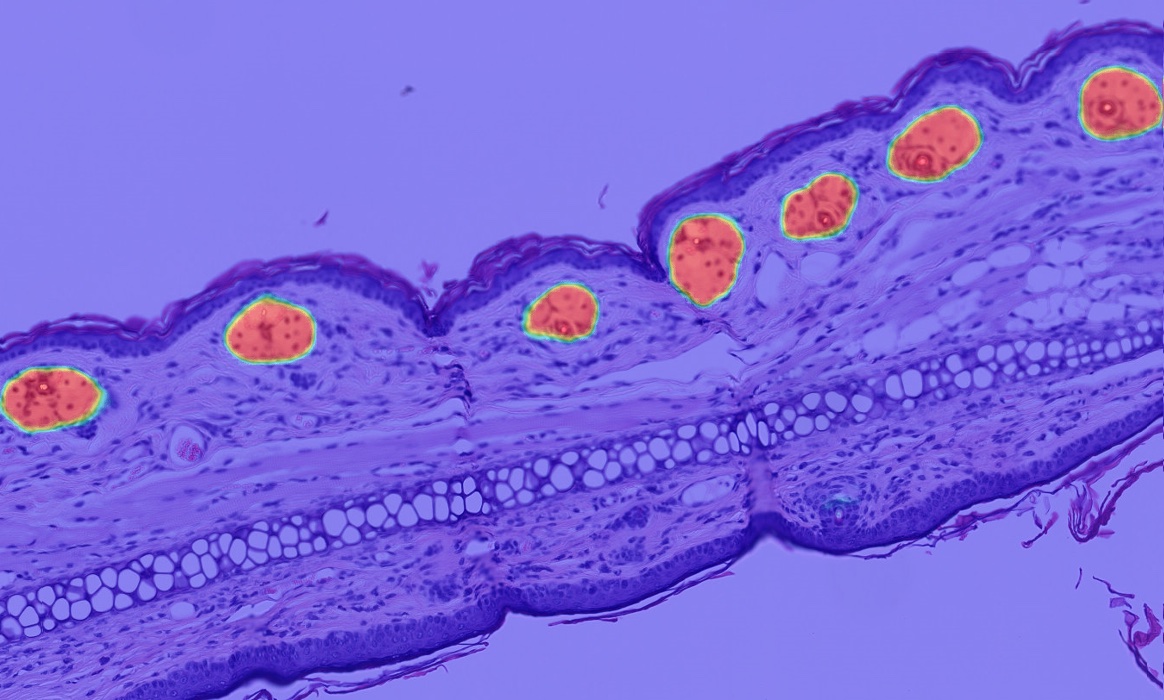
Advanced Phenotyping
Cell phenotyping can be used on any of the myriad supported image types to automatically assign cells into user defined phenotypes. Simply select a nuclear or membrane segmentation network, provide a few quick training examples, and train the network to identify phenotypes of interest.
Phenotyping is a highly effective and flexible tool to quantify cells based on morphology when biomarker information is absent and can assist with quantification and characterization of cells with complex morphologies, such as neuronal cells.
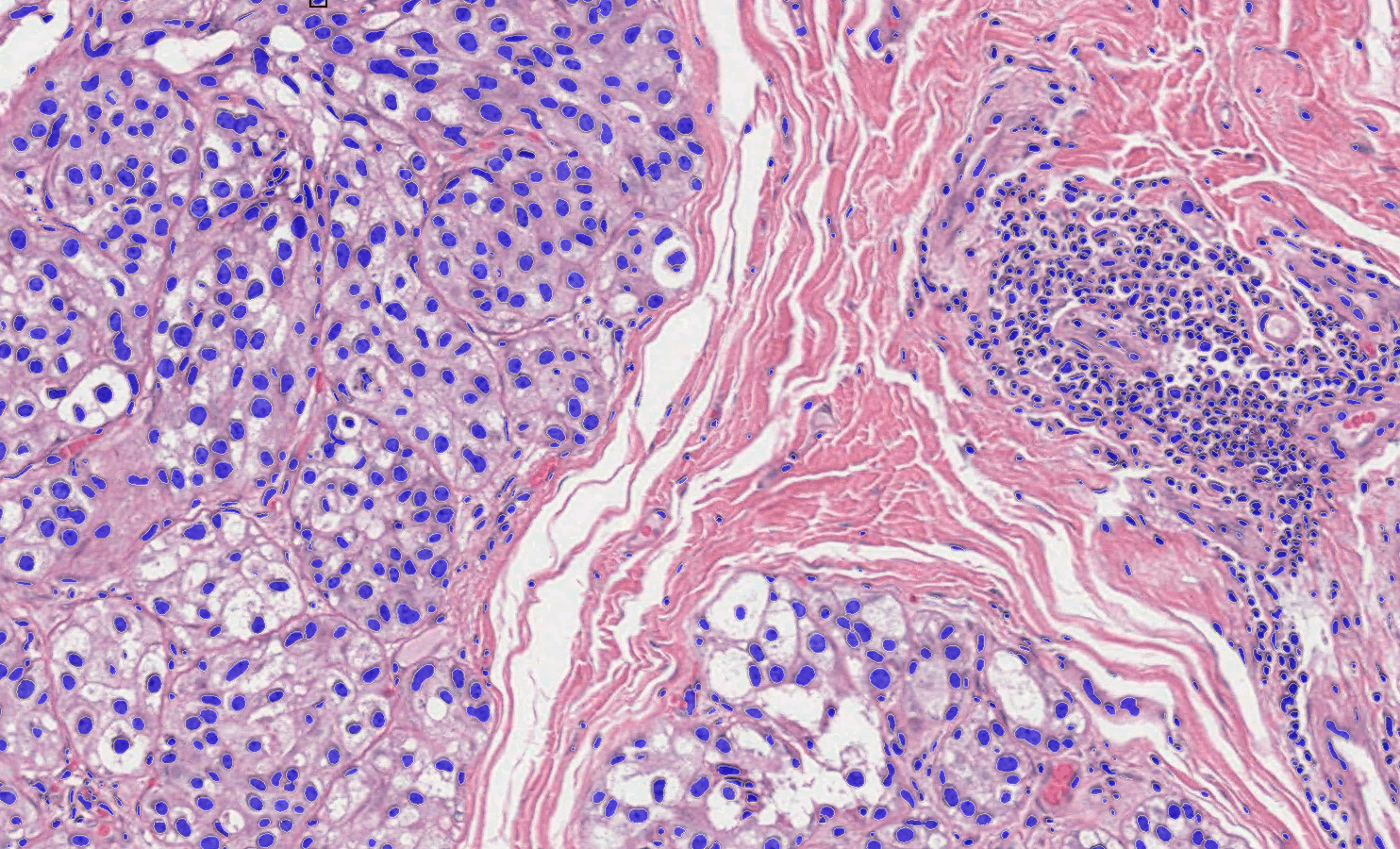
Top Notch Training and Support
At Indica Labs, we take pride in providing top-quality support. Unlimited IT support, scientific support, training, and access to our Learning Portal are included with every license at no additional cost. With support staff in the US, UK, EU, China, and Japan, you are supported in any time zone.
Wondering how easy developing your own algorithm can be without programming experience? Reach out to us to find out how intuitive HALO AI’s train-by-example interface really is.

What Our Customers Have to Say
Read independent reviews on our HALO AI Deep Learning Classifier Add-on and learn how these customers are using Indica Labs’ solutions to streamline their workflows.
Wonderful products, it is far beyond our expectations.
"Our lab both have the HALO Image Analysis System and the HALO AI, honestly speaking, the HALO system is beyond our expectations, especially the analysis process, annotation tools, the adjustable parameters, the texture and morphological recognition of nuclear and specific structure, and the after-sale care I strongly recommend you guys to choose it. it really worth the money."
lanlan Li
Panovue
Great results, can't live without this instrument!
"Scientists can achieve lots of high-quality analysis data about figures by HALO software. Many functions of HALO are very helpful to study images, such as TMA, spatial analysis, high-Plex FL, and especially HALO AI. It is easy to operate. Its interface is simple, friendly, and convenient. Once we have problems, a Halo technician is able to help us to solve them in time."
Junfeng Hao
Institute of Biophysics, Chinese Academy of Sciences
Great result. Nice tool for translational pathology research.
"We are interested in applying HALO AI in RNAscope and IHC analysis. Thanks to Yongtian ZHAO to give us wonderful training and support. We are improving and confident to apply it in the RNAscope assay development and scoring."
Fei Yang
Johnson and Johnson
Currently the best image viewer and analysis software on the market that I've used.
Nathan Aleynick
MSKCC
Platform Compatibility
HALO AI is compatible with all of the file formats that can be used in HALO and HALO Link. Yours not on the list? Email us your requirements.
File Formats:
- Non-proprietary (JPG, TIF, OME.TIFF)
- Nikon (ND2)
- 3DHistech (MRXS)
- Akoya (QPTIFF, component TIFF)
- Olympus / Evident (VSI)
- Hamamatsu (NDPI, NDPIS)
- Aperio (SVS, AFI)
- Zeiss (CZI)
- Leica (SCN, LIF)
- Ventana (BIF)
- Philips (iSyntax, i2Syntax)
- KFBIO (KFB, KFBF)
- DICOM (DCM*)
*whole-slide images
Advancing Discovery with AI: Features of HALO AI
Quickly acquire training annotations with the new AI annotation tool in HALO AI. Simply point and click your object of interest to add an annotation.
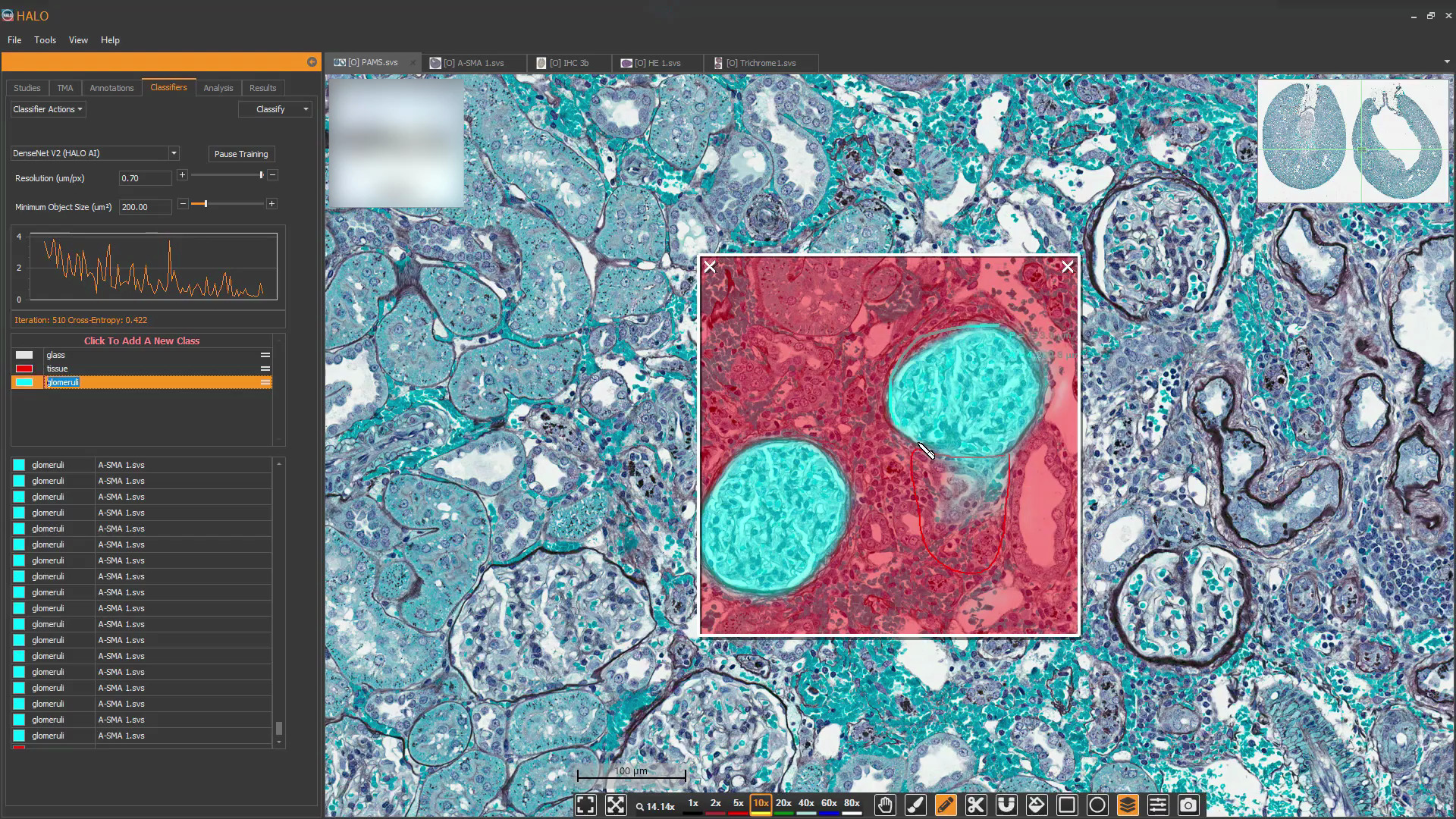
Use real-time tuning in HALO AI to watch a network as it trains in real time. Toggle the mark up on and off to evaluate performance, choose to add training data, or change parameters on-the-fly.
Once a HALO AI model is trained, a probability map can be used as an alternative output to a traditional mask to evaluate performance. Use real-time tuning to select an appropriate probability cut-off for a given class and view the output in a heatmap where blue represents the lowest probability and red the highest.
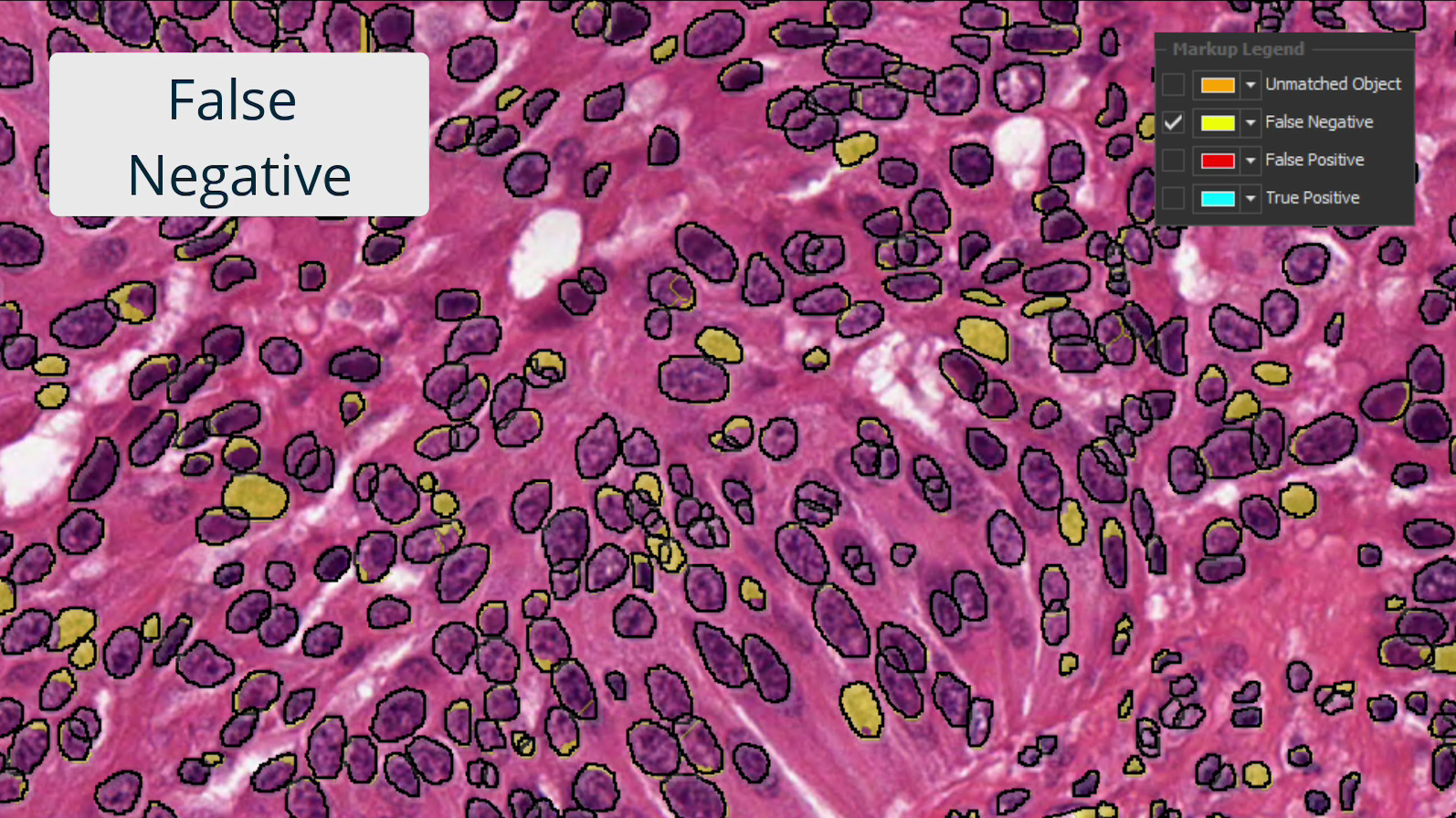
Investigate results with interactive markup images where you can toggle on and off each population of interest. Interactive markups can be combined with probability thresholding and are especially valuable in exploring validation outputs.
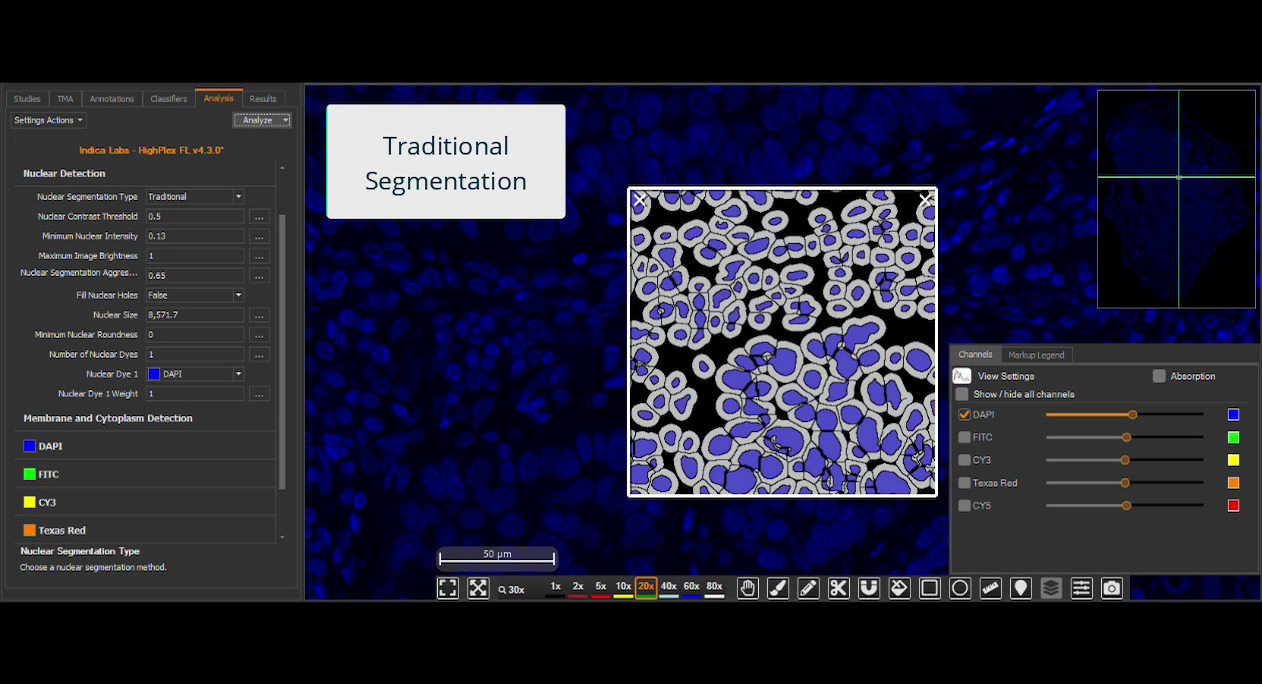
Deploy your HALO AI models in the context of HALO modules for maximum image analysis utility. Choose to utilize one or more models such as nuclear segmentation, membrane segmentation, tissue classification, cell or object phenotyping, and/or SlideQC.
Leverage the three-part Train – Validate – Test workflow to address the question: How good is my HALO AI classifier? Quantitative metrics are output based on the type of network.
Speed up your HALO AI analysis by adapting your trained network for your specific GPU. Also, you can queue training jobs based on number of iterations or time and set it to run at the end of your workday to maximize productivity and evaluate multiple model variants.
Free Image Analysis
See HALO AI in action on up to three of your images with a free analysis.
Knowledge Center
Learn more about HALO AI by exploring the tabs below. For an introduction to the capabilities of HALO AI, we recommend you check out our HALO AI white paper!


Glomeruli Quantification and Characterization Application Note
Don’t let varied stain selections hold back your digital image analysis of kidney sections! With HALO AI you can develop a tissue classifier that accurately detects and segments glomeruli in sections across a range of stains including H&E, trichrome, PAMS, and DAB with hematoxylin.
Submit the form below to view the requested document
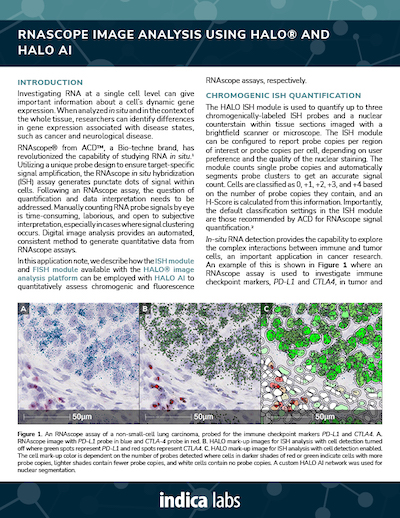
Quantify Single and Multiplexed RNAscope® Probes Across Whole Slide Tissues
Using our ISH module for brightfield and our FISH module for fluorescence, we demonstrate how to quantify ISH signal from RNAscope® assays on a per cell or per unit area basis and output an overall expression score based on ACD Bio’s recommended scoring guidelines.
Submit the form below to view the requested document

HiPlex RNAscope™ Application Note
Using our FISH module we demonstrate how to perform 12-plex RNAscope image analysis using ACD’s HiPlexv2 assay.
Submit the form below to view the requested document

Identifying Islets with HALO AI
Learn how HALO AI can detect islets of variable morphology in H&E-stained pancreatic tissue
Submit the form below to view the requested document

Camelyon 17 Application Note
CAMELYON17 challenged teams to develop automated methods for identifying and staging breast cancer metastases; read how the performance of a HALO AI classifier stood out among commercial entries!
Submit the form below to view the requested document

NCI Case Study
Download this case study to learn about considerations the National Cancer Institute made when planning to migrate its HALO, HALO AI, and HALO Link deployments to the cloud, and benefits the NCI experienced after migration, including 50% faster HALO image analysis.
Submit the form below to view the requested document

Cloud Services and AI Diagnostics Case Study
Learn how the optimized, scalable infrastructure provided by Indica Labs Cloud Services enables the rapid development and robust collaboration of our AI Diagnostics group, powering the next generation of automated AI-based decision support tools.
Submit the form below to view the requested document
Lunaphore COMET Hyperplex IF and HALO® Image Analysis eBook
Our collaborative eBook with Lunaphore shows how HALO and HALO AI, alongside the Lunaphore COMET instrument, enable a high-throughput workflow for phenotypic and spatial analysis of cells in the tumor microenvironment.
Submit the form below to view the requested document

Sequential Same Slide mIF and H&E eBook
Reveal more data on a single slide with this workflow for sequential same slide multiplex IF and H&E staining and imaging using HALO and HALO AI.
Submit the form below to view the requested document

mIF Co-Registration and Cellular Analysis eBook
Learn how HALO offers a streamlined workflow for co-registration and cellular analysis of immunofluorescence images from cyclically stained and imaged slides.
Submit the form below to view the requested document

Qualitative and Quantitative Evaluation of the TME eBook
Check out this ebook to see how AI-based nuclear segmentation and tissue classification can streamline cell phenotyping across tissue compartments and samples.
Submit the form below to view the requested document
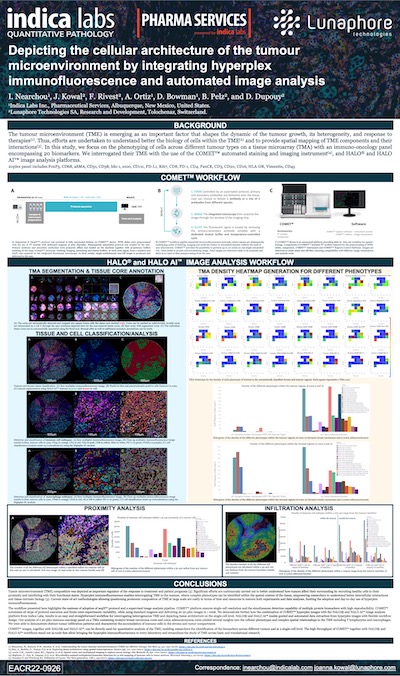
Lunaphore COMET Hyperplex IF and HALO® Image Analysis of the TME
Download our collaborative poster with Lunaphore to see how HALO and HALO AI, alongside the Lunaphore COMET instrument, enable a high throughput workflow for phenotypic and spatial analysis of cells in the tumor microenvironment.
Submit the form below to view the requested document
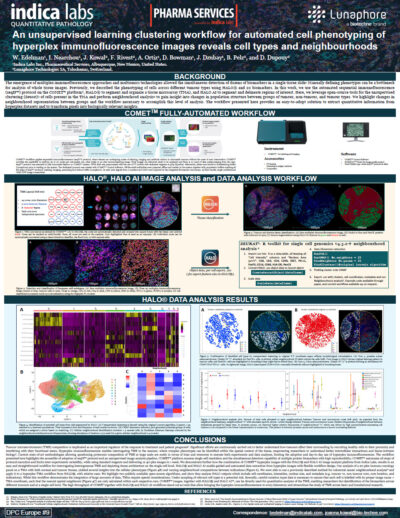
Clustering Workflow for Advanced Cell Phenotyping of Hyperplex IF Images
Submit the form below to view the requested document

Spatial Characterization of the TME with Cell DIVE Multiplexed Imaging and HALO Image Analysis
Submit the form below to view the requested document
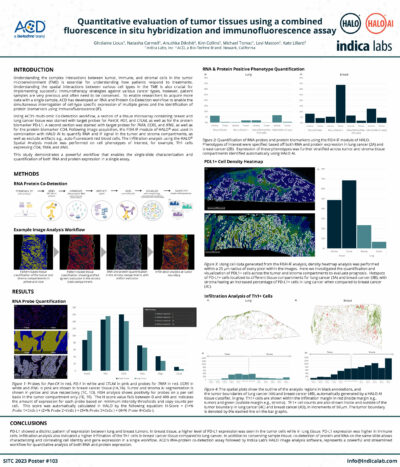
Quantitative Image Analysis of a Combined FISH-IF Assay
Submit the form below to view the requested document
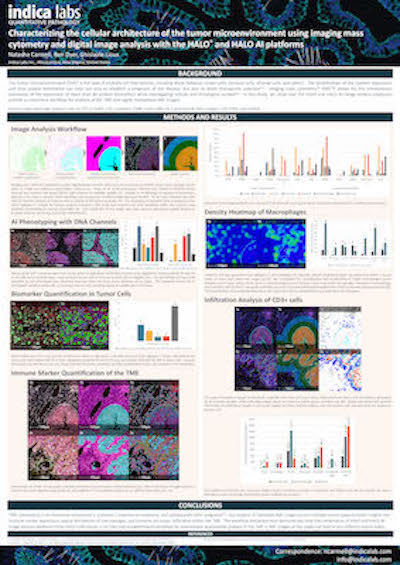
Characterizing the TME Using IMC and HALO Image Analysis
Download this poster to learn how HALO and HALO AI can be used to analyze imaging mass cytometry data.
Submit the form below to view the requested document
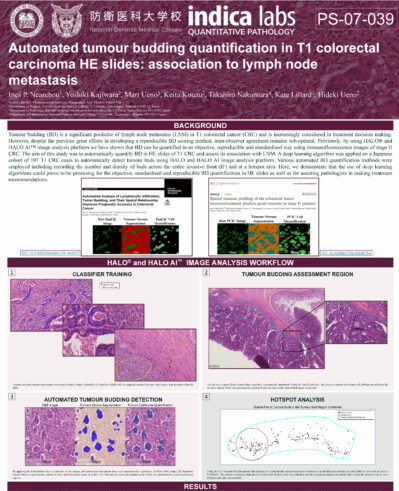
Automated Tumor Budding Quantification in Colorectal Carcinoma H&E Images
Submit the form below to view the requested document
Spatial Multiplex Profiling of Immune Markers with the RNAscope Hiplex v2 Assay
Submit the form below to view the requested document
Evaluation of the TME by High Resolution 17-plex IF and HALO Image Analysis
Submit the form below to view the requested document
Our customers are making vital discoveries in oncology, neuroscience, and diabetes research. Check out some of the research our customers are publishing using the HALO AI Deep Learning Classifier Add-on.
This study examined CD8+ cell distribution in hepatocellular carcinoma (HCC) and peritumoral liver tissue to investigate the overall survival (OS) and recurrence-free survival (RFS). It is well understood that CD8+ lymphocytes are involved in both the anti-tumor response and in...
Learn MoreEZH2 is the catalytic component of Polycomb Repressive Complex 2 (PRC2) and performs trimethylation of histone H3 at lysine 27 (H3K27me3) to silence chromatin. PRC2 is known to play different roles in different cancers and inhibitors of PRC2 histone methyltransferase...
Learn MorePredictive biomarkers to determine which non small cell lung cancer (NSCLC) patients will respond to immunotherapy are currently lacking. In this Nature Cancer publication, the authors combine computed tomography imaging, PD-L1 immunohistochemistry (IHC), and genomic data to predict responses to...
Learn More
HALO AI High Impact Publications
Review this selection of high-impact publications for examples of HALO AI applications in fields ranging from metabolism to immuno-oncology and myology.
Submit the form below to view the requested document

HALO AI White Paper
Check out this white paper for an in-depth introduction to the deep learning-based add-on to HALO, including available networks, common functions, and the “train – validate – test” Validation workflow.
Submit the form below to view the requested document

HALO Link Pharma Services White Paper
Download our white paper to learn how Indica Labs’ Pharma Services collaborates with customers and delivers HALO and HALO AI image analysis using HALO Link.
Submit the form below to view the requested document

Quantitative RNAscope™ Image Analysis Guide
From experimental design considerations to optimized setup of HALO image analysis parameters, our guide will help take your quantitative RNAscope™ image analysis to the next level.
Submit the form below to view the requested document

Pathologist-Trained Machine Learning Classifiers Quantitate Celiac Disease Features
30 May 2024 | Histologic evaluation of the mucosal changes associated with celiac disease is important for establishing an accurate diagnosis and monitoring the impact

AI-Powered Pathology with HALO®, HALO AI, and HALO Link 4.0
6 June 2024 | Join us for this 1-hour webinar to see a live demonstration of the new versions of the industry leading AI-powered HALO®
AI-based Slide Quality Control Workflows for Life Science and Clinical Applications
18 April 2024 | In this 1-hour webinar learn about SlideQC BF 3.0, an AI-based image quality control network that detects and segments most common
HALO AI 4.0 Sneak Peek
29 February 2024 | Please join us for this 1-hour webinar to learn about the exciting new features coming soon in HALO AI 4.0!

Indica Labs and Molecular Instruments Announce Partnership to Advance AI-Powered Quantitative Image Analysis with HCR™ RNA-ISH
Albuquerque, NM, and Los Angeles, CA – 5 April 2024 – Indica Labs, a leading provider of AI-powered digital pathology solutions, and Molecular Instruments® (MI),
Indica Labs Launches Comprehensive Management Service for HALO® Software in AWS Cloud-Hosted Environments
Albuquerque, NM – 8 June 2023 – Indica Labs today announces the launch of a service offering deployment and on-going support of its HALO® digital pathology
Press Release: Lunaphore and Indica Labs announce partnership to provide complete technology solution for spatial biology and image analysis
LAUSANNE, Switzerland and ALBUQUERQUE, N.M. – May 12, 2022 – 5 pm (CET) | 9 am MDT – Lunaphore, a Swiss life sciences company developing

Press Release: Indica Labs announce integration of Philips iSyntax image format across all HALO® software platforms
Albuquerque, NM– October 22, 2020 – Indica Labs, a leading provider of computational pathology software and services, is pleased to announce integration of the Philips

Looking Back: Reviewing 2023 at Indica Labs
As we welcome the new year and look back on 2023, the Indica Labs team wants to extend our heartfelt thanks to our customers and

Taking HALO® to the Cloud with Indica Labs Cloud Services
In this blog post we will discuss the basics of cloud computing, benefits and considerations of cloud vs. traditional deployments, and how our Professional
HALO®, HALO AI, and HALO Link 3.6 Features and Functionalities
In this blog post, you can learn about some of the new features in these modules, where to find the user guides, tutorial videos, and

Indica Labs’ Boston HALO® User Group Meeting
16 May 2024 | Indica Labs is pleased to announce our Boston HALO® User Group Meeting at Le Méridien Boston Cambridge on May 16 from

Indica Labs’ Frankfurt HALO® User Group Meeting
Indica Labs is pleased to announce our Frankfurt HALO® User Group Meeting at Hyatt Place Frankfurt Airport Hotel on 13 March from 12:00 – 18:00.

Indica Labs’ London HALO® User Group Meeting
6 December 2023 | Indica Labs is pleased to announce our London HALO® User Group Meeting to be held in London on 6 December 2023

Japan User Group Meeting
Indica Labs’ Japan HALO® User Group Meeting Date: 20 October 2023 Time: 10:30 – 16:00 (GMT+8) Location: Senri Room A, Senri Life Science Center, Osaka,

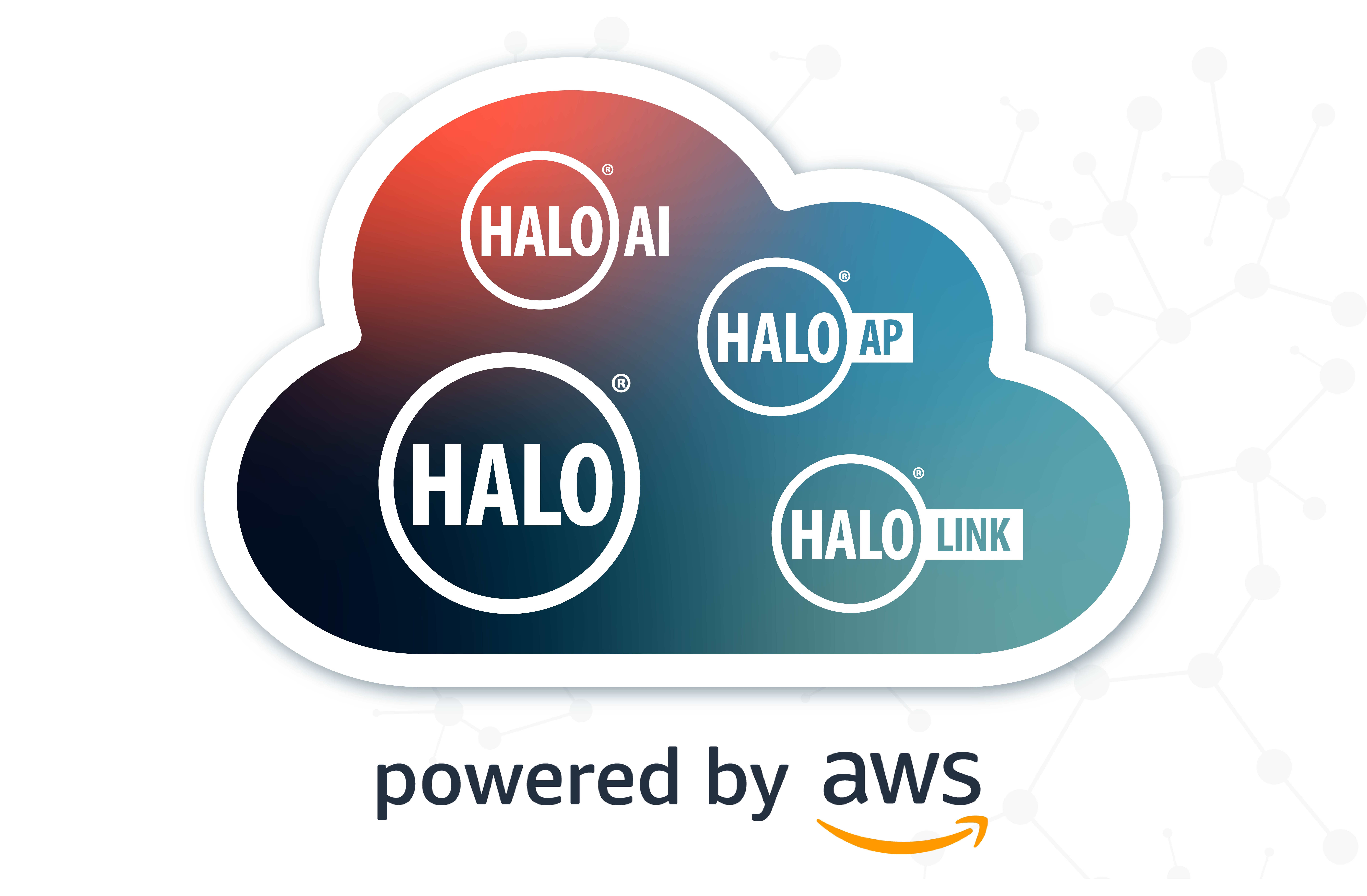
Deploy HALO® and HALO AI in a Fully Managed Cloud-Hosted Environment
Indica Labs offers optimized deployments of HALO and HALO AI software hosted in Amazon Web Services (AWS). We manage all aspects of implementation and ongoing maintenance to provide a highly performant, scalable, and secure AI-enabled HALO environment in the cloud. You maintain full ownership over your AWS account while our AWS Certified Solutions Architects manage your infrastructure and HALO software environments.
Learn more about benefits of cloud-hosted environments in our recent blog post. Or reach out today to our Cloud Services team by using the form below, to learn whether a managed cloud deployment is right for you.
Want to Learn More?
Fill out the form below to request a recorded or live demo of HALO AI or any of our other products or services.
You can also drop us an email at info@indicalab.com