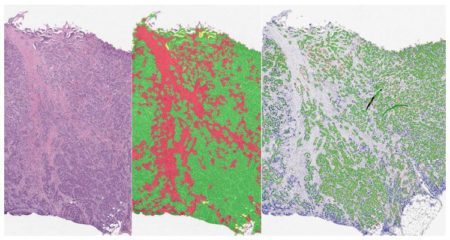
Leverage the Serial Section Analysis Add-on for HALO® to streamline your workflows for both serial section analysis and serial stain fusion analysis. For serial section analysis, the add-on enables users to create a tissue classifier based off a reference slide, such as an H&E image, and apply the resulting classification mask as regions of interest in serial section(s) stained for additional IHC markers. Serial section analysis is compatible with both brightfield and fluorescent images, and any of our HALO modules can be used for the final analysis step.
The serial stain fusion component of this add-on facilitates analysis of single slides that have been stained, stripped, and restained. In this workflow, brightfield images are deconvolved into their composite stains and converted to pseudo-fluorescent images before subsequent registration using a customizable set of configuration settings. Fluorescent images can also be registered using this workflow, just skip the deconvolution step. Lastly, the registered serial stain images can be fused into a single composite fluorescent image that can be analyzed using Highplex FL or any of our other fluorescent modules.
Want to know more? Contact us at info@indicalab.com to discuss your options with an applications scientist.

File formats supported by the HALO image analysis platform:
- Non-proprietary (JPG, TIF, OME.TIFF)
- Nikon (ND2)
- 3D Histech (MRXS)
- Akoya (QPTIFF, component TIFF)
- Olympus / Evident (VSI)
- Hamamatsu (NDPI, NDPIS)
- Aperio (SVS, AFI)
- Zeiss (CZI)
- Leica (SCN, LIF)
- Ventana (BIF)
- Philips (iSyntax, i2Syntax)
- KFBIO (KFB, KFBF)
- DICOM (DCM*)
*whole-slide images

mIF Co-Registration and Cellular Analysis eBook
Learn how HALO offers a streamlined workflow for co-registration and cellular analysis of immunofluorescence images from cyclically stained and imaged slides.
Submit the form below to view the requested document

CD38 in the Prostate Tumor Microenvironment
6 October 2021 | CD38, a druggable ectoenzyme, is involved in the generation of adenosine, which is implicated in tumor immune evasion. Its expression and

Webinar | Digital Pathology in the New Normal: Leveraging HALO® to Investigate a Global Pandemic
21 January 2021 | 8:00 AM – 9:00 AM PST | 11:00 AM – 12:00 PM EST | 4:00 PM – 5:00 PM GMT |<br

HALO® IMAGE ANALYSIS MASTERCLASS WEBINAR SERIES 2020
Summer of 2020 Indica Labs is excited to introduce our new HALO® Masterclass Webinar Series. Each masterclass webinar will offer a deep dive into a

The Use of Image Fusing in the Deployment of a 7-plex Immunofluorescent Assay
27 April 2020 | In lieu of our participation in the now cancelled 2020 AACR Conference, Indica Labs are pleased to bring you webinar presentations
Publication Spotlight
The table below includes publications that cite the Serial Section Analysis Add-on.
Your publication not on the list? Drop us an email to let us know about it!
| Title | Authors | Year | Journal | Application | HALO Modules | |
|---|---|---|---|---|---|---|
| A Cdh3-β-catenin-laminin signaling axis in a subset of breast tumor leader cells control leader cell polarization and directional collective migration | Hwang P, Mathur J, Cao Y, Almeida J, Ye J, Morikis V, Cornish D, Clarke M, Stewart S, Pathak A, Longmore G | 2023 | Developmental Cell | Oncology | Highplex FL, Registration | HALO |
| Standardized pathology screening of mature Tertiary Lymphoid Structures in cancers | Vanhersecke L, Bougouin A, Crombe A, Brunet M, Sofeu C, Parrens M, Pierron H, Bonhomme B, Lembege N, Rey C, Velasco V, Soubeyran I, Begueret H, Bessede A, Bellera C, Scoazec J-Y, Italiano A, Fridman C, Fridman W, Le Loarer F | 2023 | Laboratory Investigation | Other | Highplex FL, Registration | HALO |
| Fucosylation of HLA-DRB1 regulates CD4+ T cell-mediated anti-melanoma immunity and enhances immunotherapy efficacy | Lester D, Burton C, Gardner A, Innamarato P, Kodumudi K, Liu Q, Adhikari E, Ming Q, Williamson D, Frederick D, Sharova T, White M, Markowitz J, Cao B, Nguyen J, Johnson J, Beatty M, Mockabee-Macias A, Mercurio M, Watson G, Chen P-L, McCarthy S, MoranSegura C, Messina J, Thomas K, Darville L, Izumi V, Koomen J, Pilon-Thomas S, Ruffell B, Luca V, Haltiwanger R, Wang X, Wargo J, Boland G, Lau E | 2023 | Nature Cancer | Immuno-oncology | Highplex FL, Registration | HALO |
| CXCL9 and CXCL13 coordinately shape the immune-activated microenvironment of endometrial cancer via tertiary lymphoid structure formation | Kodama M, Nagase Y, Aimono E, Nakamura K, Takamatsu R, Abe K, Yoshimura T, Chiyoda T, Yamagami W, Aoki D, Nishihara H | 2023 | Research Square | Immuno-oncology | Registration | HALO |
| African-Ancestry Associated Gene Expression Signatures and Pathways in Triple Negative Breast Cancer, a Comparison across Women of African Descent | Martini R, Delpe P, Chu T, Arora K, Lord B, Verma A, Chen Y, Gebregzabher, Oppong J, Adjei E, Jibril A, Awuah B, Bekele M, Abebe E, Kyei I, Aitpillah F, Adinku M, Ankomah K, Osei-Bonsu E, Chitale D, Bensenhaver J, Nathanson S, Jackson L, Jiagge E, Petersen L, Proctor E, Gyan K, Gibbs L, Monojlovic Z, Kittles R, White J, Yates C, Manne U, Gardner K, Mongan N, Cheng E, Ginter P, Hoda S, Elemento O, Robine N, Sboner A, Carpten J, Newman L, Davis M | 2022 | medRxiv | Oncology | Multiplex IHC, Serial Section | |
| Multiplex Immunohistochemical Phenotyping of T Cells in Primary Prostate Cancer | Ozbek B, Ertunc O, Erickson A, Vidal I, Gomes-Alexandre C, Guner G, Hicks J, Jones T, Taube J, Sfanos K, Yegnasubramanian S, DeMarzo A | 2022 | The Prostate | Immuno-oncology | Classifier, Cytonuclear, Highplex FL, Serial Section, Registration, Figure Maker, TMA | |
| Assessment of associations between clinical and immune microenvironmental factors and tumor mutation burden in resected nonsmall cell lung cancer by applying machine learning to whole-slide images | Ono A, Terada Y, Kawata T, Serizawa M, Isaka M, Kawabata T, Imai T, Mori K, Muramatsu K, Hayashi I, Kenmotsu H, Oshima K, Urakami K, Nagashima T, Kusuhara M, Akiyama Y, Sugino T, Ohde Y, Yamaguchi K, Takahashi T T | 2020 | Cancer Medicine | Classifier, Multiplex IHC, Serial Section | ||
| Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers | Chalabi M, Fanchi LF, Dijkstra KK, Van den Berg JG, Aalbers AG, Sikorska K, Lopez-Yurda M, Grootscholten C, Beets GL, Snaebjornsson P, Maas M, Mertz J, Veninga V, Bounova G, Broeks A, Beets-Tan RG, de Wijkerslooth TR, van Lent AU, Marsman HA, Nuijten E, Kok NF, Kuiper M, Verbeek WH, Kok M, Van Leerdam ME, Schumacher TN, Voest EE, Haanen JB | 2020 | Nature Medicine | Immuno-oncology | Multiplex IHC, Serial Section | |
| Multidimensional, quantitative assessment of PD-1/PD-L1 expression in patients with Merkel cell carcinoma and association with response to pembrolizumab | Giraldo NA, Nguyen P, Engle EL, Kaunitz GJ, Cottrell TR, Berry S, Green B, Soni A, Cuda JD, Stein JE, Sunshine JC, Succaria F, Xu H, Ogurtsova A, Danilova L, Church CD, Miller NJ, Fling S, Lundgren L, Ramchurren N, Yearley JH, Lipson EJ, Cheever M, Anders RA, Nghiem PT, Topalian SL, Taube JM | 2018 | Journal for ImmunoTherapy of Cancer | Oncology, Immuno-oncology | Spatial Analysis, Serial Section |
Related HALO Modules
Quantify expression of up to five brightfield stains in any cellular compartment - membrane, nucleus or cytoplasm.
Learn MorePlot cells and objects from one or more images and perform nearest neighbor analysis, proximity analysis, and tumor infiltration analysis.
Learn MoreSeparate multiple tissue classes across a tissue using a learn-by-example approach. Can be used in conjunction with all other modules (fluorescent and brightfield) to select specific tissue classes for further analysis.
Learn MoreWant to Learn More?
Fill out the form below to request information about any of our software products.
You can also drop us an email at info@indicalab.com
Products & Services
Interested in purchasing or learning more about our products and services? Our highly trained application scientists are a couple of clicks away.
Software Maintenance & Support Coverage
Interested in purchasing an SMS plan? We would be happy to give you a quote.
Technical Support
Need technical support? Our IT specialists are here to help.