Leverage the random forest machine learning algorithm behind the Tissue Classifier Add-on for HALO® to quickly and automatically identify tissue types based on color, texture, and contextual features. This add-on features an intuitive “learn-by-example” approach and can categorize tissues within seconds after the user highlights a few distinct tissue types. The Tissue Classifier Add-on is embedded in the HALO platform and can be used in conjunction with any cell-based analysis module for brightfield or fluorescence analysis.
Try it out! Click here to initiate your free proof-of-concept HALO image analysis.
File formats supported by the HALO image analysis platform:
- Non-proprietary (JPG, TIF, OME.TIFF)
- Nikon (ND2)
- 3D Histech (MRXS)
- Akoya (QPTIFF, component TIFF)
- Olympus / Evident (VSI)
- Hamamatsu (NDPI, NDPIS)
- Aperio (SVS, AFI)
- Zeiss (CZI)
- Leica (SCN, LIF)
- Ventana (BIF)
- Philips (iSyntax, i2Syntax)
- KFBIO (KFB, KFBF)
- DICOM (DCM*)
*whole-slide images

Fusion of Spectrally Unmixed Image Tiles into Whole Slide TIFs and HALO Image Analysis
In this application note, we describe how HALO can convert spectrally unmixed image tiles into whole slide images for analysis of the tumor microenvironment.
Submit the form below to view the requested document

Spatial Analysis Application Note
In this application note, we describe two HALO analysis tools that are used to quantify the distribution of immune cells relative to tumor cells in the tumor microenvironment, Proximity Analysis and Tumor Infiltration Analysis
Submit the form below to view the requested document
HALO AI High Impact Publications
Review this selection of high-impact publications for examples of HALO AI applications in fields ranging from metabolism to immuno-oncology and myology.
Submit the form below to view the requested document

HALO AI White Paper
Check out this white paper for an in-depth introduction to the deep learning-based add-on to HALO, including available networks, common functions, and the “train – validate – test” Validation workflow.
Submit the form below to view the requested document

Quantitative RNAscope™ Image Analysis Guide
From experimental design considerations to optimized setup of HALO image analysis parameters, our guide will help take your quantitative RNAscope™ image analysis to the next level.
Submit the form below to view the requested document
HALO® High Impact Publications
Learn how HALO image analysis is leveraged in fields from immuno-oncology to neuroscience and infectious disease in this selection of high-impact publications.
Submit the form below to view the requested document

A role for HALO® in characterizing cell heterogeneity across organ systems: from the liver to the brain
12 October 2023 | Please join us for this 1-hour webinar to learn about characterizing cell heterogeneity with HALO to gain new biological insights.
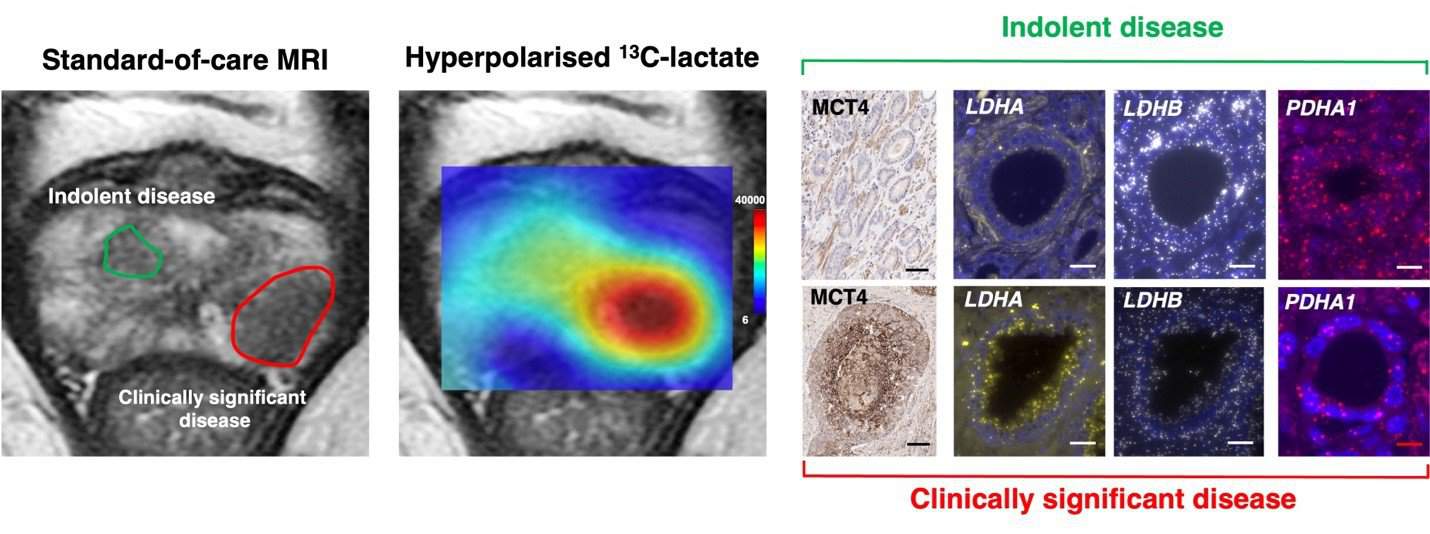
Dissecting Prostate Cancer Metabolic Compartmentalization Using Digital Pathology and Hyperpolarized MRI
6 April 2023 | Learn how HALO® image analysis coupled with hyperpolarized MRI is advancing prostate cancer research.
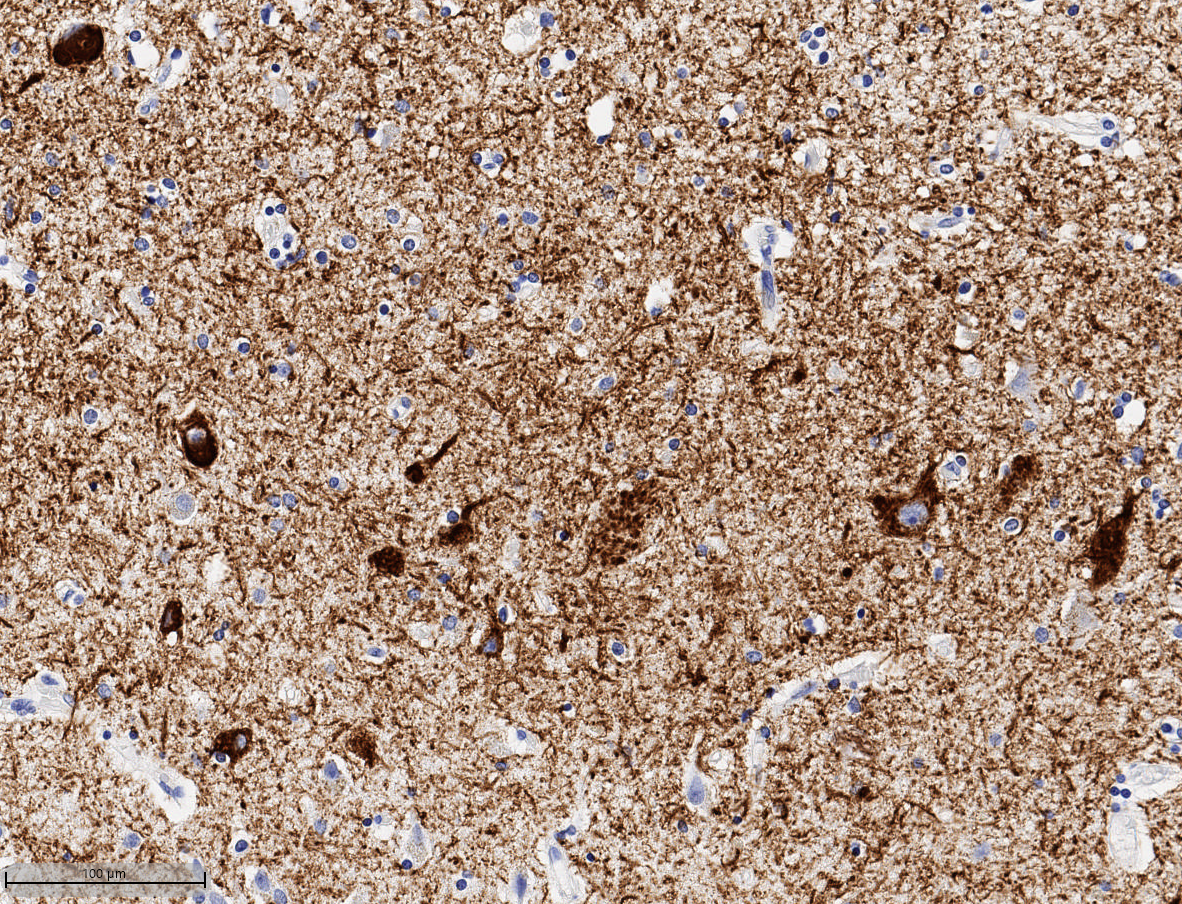
Maximizing use of HALO® and HALO AI for a comprehensive image analysis for HUMAN Brain FFPE Tissue samples in Alzheimer’s Disease
25 May 2022 | In this 60-min webinar, Learn how HALO and HALO AI are advancing neuropathology research at the UW Medicine Biorepository and Integrated
Optimizing quantitative analysis of highly multiplexed CODEX images in HALO
31 March 2022 | In this webinar, Dr. Noemi Kedei, a staff scientist at the Collaborative Protein Technology Resource core of the Center for Cancer
Publication Spotlight
The table below includes publications that cite the Tissue Classifier Add-on.
Your publication not on the list? Drop us an email to let us know about it!
| Title | Authors | Year | Journal | Application | HALO Modules | Product |
|---|---|---|---|---|---|---|
| Understanding the impact of chemotherapy on the immune landscape of high-grade serous ovarian cancer | Vanguri R, Benhamida J, Young J, Li Y, Zivanovic O, Chi D, Snyder A, Hollmann T, Mager K | 2022 | Gynecologic Oncology Reports | Oncology | Classifier, Cytonuclear, Highplex FL | HALO |
| Fasting improves therapeutic response in hepatocellular carcinoma through p53-dependent metabolic synergism | Krstic J, Reinisch I, Schindlmaier K, Galhuber M, Riahi Z, Berger N, Kupper N, Moyschewitz E, Auer M, Michenthaler H, Nossing C, Repaoli M, Ramadani-Muja J, Usluer S, Stryeck S, Pichler M, Rinner B, Deutsch A, Reinisch A, Madl T, Chiozzi R, Heck A, Huch M, Malli R, Prokesch | 2022 | Science Advances | Oncology | Classifier | HALO |
| Therapeutic treatment with the anti-inflammatory drug candidate MW151 may partially reduce memory impairment and normalizes hippocampal metabolic markers in a mouse model of comorbid amyloid and vascular pathology | Braun D, Powell D, McLouth C, Roy S, Watterson D, Van Eldik | 2022 | PLOS One | Neruoscience | Classifier, Area Quantification | HALO |
| A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo | Kofink C, Trainor N, Mair B, Wrhrle S, Wurm M, Mischerikow N, Bader G, Rumpel K, Gerstberger T, Cui Y, Greb P, Garavel G, Scharnweber M, Fuchs J, Gremel G, Chette P, Hopf S, Budano N, Rinnenthal J, Gmaschitz G, Diers E, McLennan R, Roy M, Whitworth C, Vetma V, Mayer M, Koegl M, Ciulli A, Weinstabl H, Farnaby W | 2022 | Biological and Medicinal Chemistry | Classifier, Cytonuclear, Multiplex IHC | HALO | |
| Pancreas resident macrophage-induced fibrosis has divergent roles in pancreas inflammatory injury and PDAC | Baer J, Zuo C, Kang L, Alarcon de la Lastra A, Bocherding N, Knolhoff B, Bogner S, Zhu Y, Lewis M, Zhang N, Kim K, Fields R, Mills J, Ding L, Randolph G, DeNardo D | 2022 | bioRxiv | Fibrosis | Classifier, Area Quantification, Cytonuclear, Multiplex IHC, Highplex FL, Figure Maker | HALO |
| Fatal Neurodissemination and SARS-CoV-2 Tropism in K18-hACE2 Mice Is Only Partially Dependent on hACE2 Expression | Carossino M, Kenney D, O'Connell A, Montarnaro P, Tseng A, Gertje H, Grosz K, Ericsson M, Huber B, Kurnick S, Subramaniam S, Kirkland T, Walker J, Francis K, Klose A, Paragas N, Bosmann M, Saeed M, Balasuriya U, Douam F, Crossland N | 2022 | Viruses | Infectious Disease | Classifier, Area Quantification, Highplex FL | HALO |
| Multiplex Immunohistochemical Phenotyping of T Cells in Primary Prostate Cancer | Ozbek B, Ertunc O, Erickson A, Vidal I, Gomes-Alexandre C, Guner G, Hicks J, Jones T, Taube J, Sfanos K, Yegnasubramanian S, DeMarzo A | 2022 | The Prostate | Immuno-oncology | Classifier, Cytonuclear, Highplex FL, Serial Section, Registration, Figure Maker, TMA | HALO |
| Genome-wide mapping of oncogenic pathways and genetic modifiers of chemotherapy using a high-risk hepatoblastoma genetic model | Fang J, Singh S, Natarajan S, Tillman H, Abuzaid A, Durbin A, Lee H, Wu Q, Steele J, Connelly J, Cheng C, Jin H, Chen W, Fan Y, Pruett-Miller S, Rehg J, Emmons J, Cairo S, Wang R, Glazer E, Murphy A, Chen T, Davidoff A, Easton J, Chen X, Yang J | 2022 | Research Square | Oncology | Classifier | HALO |
| Loss of ubiquitin-specific peptidase 18 destabilizes 14-3-3? protein and represses lung cancer metastasis | Chen Z, Zheng L, Chen Y, Liu X, Kawakami M, Mustachi L, Roszik J, Ferry-Galow K, Parchment R, Liu X, Andresson T, Duncan G, Kurie J, Rodriguez-Canales J, Liu X, Dmitrovsky E | 2022 | Cancer Biology & Therapy | Oncology | Classifier, Cytonuclear | HALO |
| Therapeutic impact of BET inhibitor BI 894999 treatment: backtranslation from the clinic | Tontsch-Grunt U, Traexler P, Baum A, Musa H, Marzin K, Wang S, Trapani F, Engelhardt H, Solca F | 2022 | British Journal of Cancer | Oncology | Classifier, Cytonuclear, Multiplex IHC | HALO |
| 3íUTR-directed, kinase proximal mRNA decay inhibits C/EBP? phosphorylation/activation to suppress senescence in tumor cells | Salotti J, Karim B, Misra S, Hu L, Basu S, Martin N, Luke B, Saylor K, Andresson T, Scheiblin D, Lockett S, Tessarollo L, Johnson P | 2022 | bioRxiv | Oncology | Classifier, Cytonuclear | HALO, HALO AI |
| Orchestrating the Dermal/Epidermal Tissue Ratio during Wound Healing by Controlling the Moisture Content | Tuca A, de Mattos I, Funk M, Winter R, Palackic A, Groeber-Becker F, Kruse D, Kukla F, Lemarchand T, Kamolz L | 2022 | Biomedicines | Other | Classifier | HALO |
| A lncRNA signature associated with tumor immune heterogeneity predicts distant metastasis in locoregionally advanced nasopharyngeal carcinoma | Liang Y, Zhang Y, Tan X, Qiao H, Liu S, Tang L, Mao Y, Chen L, Li W, Zhou G, Zhao Y, Li J, Li Q, Huang S, Gong S, Zheng Z, Li Z, Sun Y, Jiang W, Ma J, Li Y, Liu N | 2022 | Nature Communications | Immuno-oncology | Classifier, Multiplex IHC, Registration | HALO |
| Effects of Extracorporeal Shockwave Therapy on Functional Recovery and Circulating miR-375 and miR-382-5p after Subacute and Chronic Spinal Cord Contusion Injury in Rats | Ashmwe M, Posa K, Ruhrnossl A, Heinzel J, Heimel P, Mock M, Schadl B, Keibl C, Couillard-Despres S, Redl H, Mittermayr R, Hercher D | 2022 | Biomedicines | Neuroscience, Other | Classifier, Multiplex IHC | HALO |
| The combination of gene hyperamplification and PD-L1 expression as a biomarker for the clinical benefit of tislelizumab in gastric/gastroesophageal junction adenocarcinoma | Lu Z, Yang S, Luo X, Shi Y, Lee J, Deva S, Liu T, Chao Y, Zhang Y, Huang R, Xu Y, Shen Z, Shen L | 2022 | Gastric Cancer | Immuno-oncology | Classifier, Multiplex IHC | HALO |
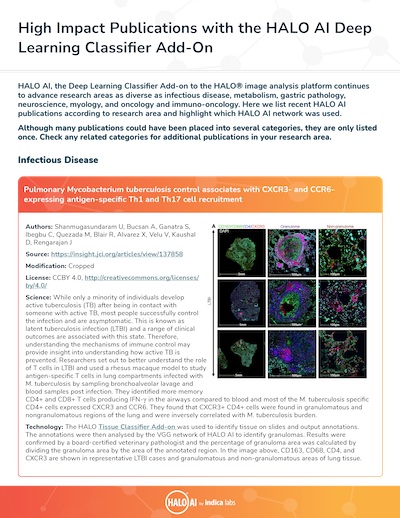
| Isoniazid and Rifapentine Treatment effectively reduces persistent M. tuberculosis infection 2 in macaque lungs | Sharan R, Ganatra S, Singh D, Cole J, Foreman T, Thippeshappa R, Peloquin C, Shivanna V, Gonzalez O, Day C, Gandhi N, Dick E, Hall-Ursone S, Mehra S, Schlesinger L, Rengarajan J, Kaushal D | 2022 | Journal of Clinical Investigation | Infectious Disease | Classifier | HALO |
| Prostate cancer androgen receptor activity dictates efficacy of Bipolar Androgen Therapy | Sena L, Kumar R, Sanin D, Thompson E, Rosen D, Dalrymple S, Antony L, Yang Y, Comes-Alexandre C, Hicks J, Jones T, Bowers K, Eskra J, Meyers J, Gupta A, Skaist A, Yegnasubramanian S, Luo J, Brennen W, Kachhap S, Antonarakis E, De Marzo A, Isaacs J, Markowski M, Denmeade S | 2022 | medRxiv | Oncology | Classifier, Cytonuclear, Multiplex IHC, Registration | HALO |
| Senescence-Associated Molecules and Tumor-Immune-Interactions as Prognostic Biomarkers in Colorectal Cancer | Kellers F, Fernandez A, Konykiewitz B, Schindeldecker M, Tagscherer K, Heintz A, Jesinghaus M, Roth W, Foersch S | 2022 | Frontiers in Medicine | Oncology | Classifier, Multiplex IHC, Spatial Analysis, Registration, TMA | HALO |
| A survey on artificial intelligence in histopathology image analysis | Abdelsamea M, Zidan U, Senousy Z, Gaber M, Rakha E, Ilyas M | 2022 | WIREs Data Mining and Knowledge Discovery | Review | Classifier, Cytonuclear, Figure Maker, TMA | HALO |
| Automated recognition of glomerular lesions in the kidneys of mice by using deep learning | Akatsuka A, Horai Y, Akatsuka A | 2022 | Journal of Pathology Informatics | Other | Classifier | HALO, HALO AI |
| MYC sensitises cells to apoptosis by driving energetic demand. | Edwards-Hicks J, Su H, Mangolini M, Yoneten K, Wills J, Rodriguez-Blanco G, Young C, Cho K, Barker H, Muir M, Guerrieri A, Li X, White R, Manasterski P, Mandrou E, Wills K, Chen J, Abraham E, Sateri K, Qian B, Bankhead P, Arends M, Gammoh N, von Kriegsheim A, Patti G, Sims A, Acosta J, Brunton V, Kranc K, Christophorou M, Pearce E, Ringshausen I, Finch A | 2022 | Nature Communications | Oncology, Metabolism | Classifier | HALO |
| Glycosphingolipids are mediators of cancer plasticity through independent signaling pathways | Cumin C, Huang Y, Rossdam C, Ruoff F, Cespedes S, Liang C, Lombardo F, Coelho R, Rimmer N, Konantz M, Lopez M, Alam S, Schmidt A, Calabrese D, Fedier A, Vlajnic T, Itzstein M, Templin M, Buettner F, Everest-Dass A, Heinzelmann-Schwarz V, Jacob F | 2022 | Cell reports | Oncology | Classifier, Multiplex IHC | HALO |
| Unveiling the tumor immune microenvironment of organ-specific melanoma metastatic sites | Conway J, Rawson R, Lo S, Ahmed T, Veregara I, Gide T, Attrill G, Carlino M, Saw R, Thompson J, Spillane A, Shannon K, Shivalingam B, Menzies A, Wilmott J, Long G, Scolyer R, da Silva I | 2022 | Journal for ImmunoTherapy of Cancer | Oncology | Classifier, Spatial Analysis, Highplex FL | HALO |
| Proliferative tumor infiltrating lymphocytes abundance within microenvironment impact clinical outcome in cutaneous B-cell lymphomas | Menguy S, Prochazkova-Carolotti M, Azzi-Martin L, Ferte T, Bresson-Bepoldin L, Rey C, Vergier B, Merlio J, Beylot-Barry M, Pham-Ledard A | 2022 | Journal of Investigative Dermatology | Immuno-oncology | Classifier, Highplex FL | HALO |
| Integrated pathological analysis to develop a Gal-9 based immune survival stratification to predict the outcome of lung large cell neuroendocrine carcinoma and its usefulness in immunotherapy | Che Y, Luo Z, Cao Y, Wang J, Xue Q, Sun N, He J | 2022 | International Journal of Biological Sciences | Oncology | Classifier, Multiplex IHC | HALO |
| PD-L1 predicts chemotherapy resistance in large-cell neuroendocrine carcinoma | Che Y, Luo Z, Cao Y, Sun N, Xue Q, He J | 2022 | British Journal of Surgery | Oncology | Classifier, Multiplex IHC | HALO |
| Systemic Nos2 Depletion and Cox inhibition limits TNBC disease progression and alters lymphoid cell spatial orientation and density | Somasundaram V, Ridnour L, Cheng R, Walke A, Kedei N, Bhattacharyya D, Wink A, Edmondson E, Butcher D, Warner A, Dorsey T, Scheiblin D, Heinz W, Bryant R, Kinders R, Lipkowitz S, Wong S, Pore M, Hewitt S, McVicar D, Anderson S, Chang J, Glynn S, Ambs S, Lockett S, Wink D | 2022 | Redox Biology | Oncology | Classifer, ISH/FISH, Spatial Analysis, Registration, ISH-IHC/FISH-IF | HALO |
| Method To Visualize the Intratumor Distribution and Impact of Gemcitabine in Pancreatic Ductal Adenocarcinoma by Multimodal Imaging | Strittmatter N, Richards F, Race A, Ling S, Sutton D, Nilsson A, Wallez Y, Barnes J, Maglennon G, Gopinathan A, Brais R, Wong E, Serra M, Atkinson J, Smith A, Wilson J, Hamm G, Johnson T, Dunlop C, Kaistha B, Bunch J, Sansom O, Takats Z, Andren P, Lau A, Barry S, Goodwin R, Jodrell D | 2022 | Analytical Chemistry | Oncology | Classifier, Highplex FL | HALO, HALO AI |
| Bone quality and composition are influenced by egg production, layer line, and estradiol-17ß in laying hens | Eusemann B, Ulrich R, Sacnhez-Rodriquez E, Benavides-Reyes C, Dominguez-Gasca, Rodriguez-Navarro A, Petow S | 2022 | Avian Pathology | Other | Classifier, Multiplex IHC | HALO |
| Tissue-resident FOLR2+ macrophages associate with CD8+ T cell infiltration in human breast cancer | Ramos R, Missolo-Koussou Y, Gerber-Ferder Y, Bromley C, Bugatti M, Nunez N, Boari J, Richer W, Menger L, Denizeau J, Sedlik C, Caudana P, Kotsias F, Niborski L, Viel S, Bohec M, Lameiras S, Baulande S, Lesage L, Nicolas A, Meseure D, Vincent-Salomon A, Reyal F, Dutertre C, Ginhoux F, Vimeux L, Donnadieu E, Buttard B, Galon J, Zelenay S, Vermi W, Guermonprez P, Piaggio E, Helft J | 2022 | Cell | Immuno-oncology | Classifier, Spatial Analysis, Highplex FL | HALO |
| Inflammatory Monocytes Promote Granuloma-Mediated Control of Persistent Salmonella Infection | Bettke J, Tam J, Montoya V, Butler B, vander Velden A | 2022 | Infection and Immunity | Infectious Disease | Classifier, Area Quantification | HALO |
| Reduced SARS-CoV-2 disease outcomes in Syrian hamsters receiving immune sera: Quantitative image analysis in pathologic assessments | Pieda-Mora C, Robinson S, Tostanoski L, Dayao D, Chandrashekar A, Bauer K, Wrijil L, Ducat S, Hayes T, Yu J, Bondzie E, McMahan K, Sellers D, Giffin V, Hope D, Nampanya F, Mercado N, Kar S, Andersen H, Tzipori S, Barouch D, Martinot A | 2022 | Veterinary Pathology | Infectious Disease, Other | Classifier, Multiplex IHC | HALO |
| Integrative Analysis of Bioinformatics and Machine Learning Algorithms Identifies a Novel Diagnostic Model Based on Costimulatory Molecule for Predicting Immune Microenvironment Status in Lung Adenocarcinoma | Zhai W, Duan F, Wang Y, Wang J, Zhao Z, Lin Y, Rao B, Chen S, Zheng L, Long H | 2022 | The American Journal of Pathology | Oncology | Classifier, Multiplex IHC | HALO |
| NKG2A and HLA-E define an alternative immune checkpoint axis in bladder cancer | Salome B, Sfakianos J, Ranti D, Daza J, Bieber C, Charap A, Hammer C, Banchereau R, Farkas A, Ruan D, Izadmehr S, Geanon D, Kelly G, Real R, Lee B, Beaumont K, Shroff S, Wang Y, Wang Y, Thin T, Garcia-Barros M, Hegewisch-Solloa E, Mace E, Wang L, O'Donnell T, Chowell D, Fernandez-Rodriguez R, Skobe M, Taylor N, Kim-Schulze S, Sebra R, Palmer D, Clancy-Thompson E, Hammond S, Kamphorst A, Malmberg K, Marcenaro E, Romero P, Brody R, Viard M, Yuki Y, Martin M, Carrington M, Mehrazin R, Wilund P, Mellman I, Mariathasan S, Zhu J, Galsky M, Bhardwaj N, Horowitz A | 2022 | Cancer Cell | Oncology, Immuno-oncology | Classifier, Multiplex IHC | HALO, HALO AI |
| Chronic rhinosinusitis patients display an aberrant immune cell localisation with enhanced S. aureus biofilm metabolic activity and biomass | Shaghayegh G, Cooksley C, Bouras G, Panchatcharam B, Idrizi R, Jana M, Ellis S, Psaltis A, Wormald P, Vreugde S | 2022 | Journal of Allergy and Clinical Immunology | Immunology, Infectious Disease | Classifier, Spatial Analysis, Highplex FL | HALO |
| Quantitative Dual-Energy Computed Tomography Image Guidance for Thermochemical Ablation: in vivo Results in the Rabbit VX2 Model | Thompson E, Fowlkes N, Jacobsen M, Layman R, Cressman E | 2022 | Journal of Vascular and Interventional Radiology | Oncology | Classifier | HALO |
| Innate immune receptor C5aR1 regulates cancer cell fate and can be targeted to improve radiotherapy in tumours with immunosuppressive microenvironments | Beach C, MacLean D, Majorova D, Melemenidis S, Nambiar D, Kim R, Valbuena G, Guglietta S, Krieg C, Darvish D, Suwa T, Eason A, Domingo E, Moon E, Jiang D, Jiang Y, Koong A, Woodruff T, Graves E, Maughan T, Buczacki S, Stucki M, Le Q-T, Leedham S, Giaccia A, Olcina M | 2023 | bioRxiv | Immuno-oncology | Classifier, Area Quantification, Highplex FL | HALO |
| NADK-mediated de novo NADP(H) synthesis is a metabolic adaptation essential for breast cancer metastasis | Ilter D, Drapela S, Schild T, Ward N, Adhikari E, Low V, Asara J, Oskarsson T, Lau E, DeNicola G, McReynolds M, Gomes A | 2023 | Redox Biology | Oncology, Metabolism | Classifier, Highplex FL, TMA | HALO |
| MYC-driven increases in mitochondrial DNA copy number occur early and persist throughout prostatic cancer progression | Chen J, Zheng Q, Hicks J, Trabzonlu L, Ozbek B, Jones T, Vaghasia A, Larman T, Wang R, Markowski M, Denmeade S, Pienta K, Hruban R, Antonaraskis E, Gupta A, Dang C, Yegnasubramanian S, De Marzo A | 2023 | bioRxiv | Oncology | Classifier, Area Quantification | HALO |
| PD-1+CD8+ T Cells Proximal to PD-L1+CD68+ Macrophages Are Associated with Poor Prognosis in Pancreatic Ductal Adenocarcinoma Patients | Yang X, Wang G, Song Y, Zhuang T, Li Y, Xie Y, Fei X, Zhao Y, Xu D, Hu Y | 2023 | Cancers | Oncology, Immuno-oncology | Classifier, Spatial Analysis, Highplex FL, Registration, TMA | HALO |
| Targeting XBP1 mRNA splicing sensitizes glioblastoma to chemotherapy | Dowdell A, Marsland M, Faulkner S, Gedye C, Lynam J, Griffin C, Marsland J, Jiang C, Hondermarck H | 2023 | FASEB BioAdvances | Oncology | Classifier, Cytonuclear | HALO |
| Spatial multi-omics revealed the impact of tumor ecosystem heterogeneity on immunotherapy efficacy in patients with advanced non-small cell lung cancer treated with bispecific antibody | Song X, Xiong A, Wu F, Li X, Wang J, Jiang T, Chen P, Zhang X, Zhao Z, Liu H, Cheng L, Zhao C, Wang Z, Pan C, Cui X, Xu T, Luo H, Zhou C | 2023 | Journal for ImmunoTherapy of Cancer | Immuno-oncology | Classifier, Highplex FL | HALO |
| Myeloid-specific deletion of activating transcription factor 6 alpha increases CD11b+ macrophage subpopulations and aggravates lung fibrosis | Mekhael O, Revill S, Hayat A, Cass S, MacDonald K, Vierhout M, Ayoub A, Reihani A, Padwal M, Imani J, Ayaub E, Yousof T, Dvorkin-Gheva A, Rullo A, Hirota J, Richards C, Bridgewater D, Stämpfli M, Hambly N, Naqvi A, Kolb M, Ask K | 2023 | Immunology and Cell Biology | Immunology, Fibrosis | Classifier, Area Quantification, FISH, Multiplex IHC | HALO |
| Cellular mechanisms of heterogeneity in NF2-mutant schwannoma | Chiasson-MacKenzie C, Vitte J, Liu C, Wright E, Flynn E, Stott S, Giovannini M, McClatchey A | 2023 | Nature Communications | Oncology | Classifier, Multiplex IHC, Spatial Analysis, FISH-IF | HALO |
| YBX1 integration of oncogenic PI3K/mTOR signalling regulates the fitness of malignant epithelial cells | Bai Y, Gotz C, Chincarini G, Zhao Z, Slaney C, Boath J, Furic L, Angel C, Jane S, Phillips W, Stacker S, Farah C, Darido C | 2023 | Nature Communications | Oncology | Classifier, Multiplex IHC | HALO |
| Prognostic value of tumor immune microenvironment factors in patients with stage I lung adenocarcinoma | Xue Q, Wang Y, Zheng Q, Chen L, Lin Y, Jin Y, Shen X, Li Y | 2023 | American Journal of Cancer Research | Oncology | Classifier, Multiplex IHC, Highplex FL | HALO |
| Industrialized, Artificial Intelligence-guided Laser Microdissection for Microscaled Proteomic Analysis of the Tumor Microenvironment | Mitchell D, Hunt A, Conrads K, Hood B, Makohon-Moore S, Rojas C, Maxwell G, Bateman N, Conrads T | 2022 | JOVE | Other | Classifier | HALO |
| Host, reproductive, and lifestyle factors in relation to quantitative histologic metrics of the normal breast | Abubakar M, Klein A, Fan S, Lawrence S, Mutreja K, Henry J, Pfeiffer R, Duggan M, Gierach G | 2023 | Research Square | Oncology | Classifier | HALO, HALO Link |
| Outdoor air pollution and histologic composition of normal breast tissue | Ish J, Abubakar M, Fan S, Jones R, Niehoff N, Henry J, Gierach G, White A | 2023 | Environment International | Oncology | Classifier | HALO |
| SRF617 Is a Potent Inhibitor of CD39 with Immunomodulatory and Antitumor Properties | Warren M, Matissek S, Rausch M, Panduro M, Hall R, Dulak A, Brennan D, Das Yekkirala S, Koseoglu S, Masia R, Yang Y, Reddy N, Prenovitz R, Strand J, Zaidi T, Devereaux E, Jacoberger Foissac C, Stagg J, Lee B, Holland P, Palombella V, Lake A | 2023 | ImmunoHorizons | Immuno-oncology | Classifier, Area Quantification, Multiplex IHC | HALO |
| Sequential pembrolizumab cooperates with platinum/5FU to remodel the tumor immune microenvironment in advanced gastric cancer: A phase II chemoimmunotherapy trial | Klempner S, Lee J, Mehta A, An M, Min B, Heo Y, Parikh M, Bi L, Cristescu R, Lee H, Kim T, Lee S, Moon J, Park R, Strickland M, Park W, Kang W, Kim K, Kim S | 2023 | Research Square | Immuno-oncology | Classifier, Highplex FL | HALO |
| NLRP12-PANoptosome activates PANoptosis and pathology in response to heme and PAMPs | Sundaram B, Pandian N, Mall R, Wang Y, Sarkar R, Kim H, Malireddi R, Karki R, Janke L, Vogel P, Kanneganti T | 2023 | Cell | Immunology | Classifier | HALO |
| Epitope Lability of Phosphorylated Biomarkers of the DNA Damage Response Pathway Results in Increased Vulnerability to Effects of Delayed or Incomplete Formalin Fixation | Wiseman E, Moss J, Atkinson J, Baakza H, Hayes E, Willis S, Waring P, Canales J, Jones G | 2023 | Journal of Histochemistry & Cytochemistry | Other | Classifier, Cytonuclear, Spatial Analysis | HALO |
| Defective extracellular matrix remodeling in brown adipose tissue is associated with fibro-inflammation and reduced diet-induced thermogenesis | Pellegrinelli V, Figueroa-Juárez E, Samuelson I, U-Din M, Rodriguez-Fdez S, Virtue S, Leggat J, Çubuk C, Peirce V, Niemi T, Campbell M, Rodriguez-Cuenca S, Dopazo Blázquez J, Carobbio S, Virtanen K, Vidal-Puig A | 2023 | Cell Reports | Metabolism | Classifier, Area Quantification, Vacuole | HALO |
| Modifying antibody-FcRn interactions to increase the transport of antibodies through the blood-brain barrier | Tien J, Leonoudakis D, Petrova R, Trinh V, Taura T, Sengupta D, Jo L, Sho A, Yun Y, Doan E, Jamin A, Hallak H, Wilson D, Stratton J | 2023 | mABS | Neuroscience | Classifier, Area Quantification, Object Colocalization | HALO |
| The antiviral effects of a MEK1/2 inhibitor promote tumor regression in a preclinical model of human papillomavirus infection-induced tumorigenesis | Luna A, Young J, Sterk R, Bondu V, Schultz F, Kusewitt D, Kang H, Ozbun M | 2023 | Antiviral Research | Oncology, Infectious Disease | Classifier, Area Quantification, Cytonuclear | HALO |
| Genome-wide mapping of cancer dependency genes and genetic modifiers of chemotherapy in high-risk hepatoblastoma | Fang J, Singh S, Cheng C, Natarajan S, Sheppard H, Abu-Zaid A, Durbin A, Lee H, Wu Q, Steele J, Connelly J, Jin H, Chen W, Fan Y, Pruett-Miller S, Rehg J, Koo S, Santiago T, Emmons J, Cairo S, Wang R, Glazer E, Murphy A, Chen T, Davidoff A, Armengol C, Easton J, Chen X, Yang J | 2023 | Nature Communications | Oncology | Classifier | HALO |
| Poor outcome in postpartum breast cancer patients is associated with distinct molecular and immunological features | Lefrère H, Moore K, Floris G, Sanders J, Seignette I, Bismeijer T, Peters D, Broeks A, Hooijberg E, Van Calsteren K, Neven P, Warner E, Peccatori F, Loibl S, Maggen C, Han S, Jerzak K, Annibali D, Lambrechts D, de Visser K, Wessels L, Lenaerts L, Amant F | 2023 | Clinical Cancer Research | Oncology | Classifier, Highplex FL | HALO, HALO AI |
| Differential expression of CD8 defines phenotypically distinct cytotoxic T cells in cancer and multiple sclerosis | Burkard T, Herrero San Juan M, Dreis C, Kiprina A, Namgaladze D, Siebenbrodt K, Luger S, Foerch C, Pfeilschifter J, Weigert A, Radeke H | 2023 | Clinical and Translational Medicine | Oncology, immunology | Classifier, Spatial Analysis, Highplex FL, TMA | HALO |
| Low molecular weight heparin synergistically enhances the efficacy of adoptive and anti-PD-1-based immunotherapy by increasing lymphocyte infiltration in colorectal cancer | Quan Y, He J, Zou Q, Zhang L, Sun Q, Huang H, Li W, Xie K, Wie F | 2023 | Journal for ImmunoTherapy of Cancer | Immuno-oncology | Classifier, Multiplex IHC, Highplex FL | HALO |
| Investigating the Effects of Stereotactic Body Radiation Therapy on Pancreatic Tumor Hypoxia and Microvasculature in an Orthotopic Mouse Model using Intravital Fluorescence Microscopy | Samuel T, Rapic S, Lindsay P, DaCosta RS | 2023 | Research Square | Oncology | Classifier, Area Quantification | HALO |
| HER2-Selective and Reversible Tyrosine Kinase Inhibitor Tucatinib Potentiates the Activity of T-DM1 in Preclinical Models of HER2-positive Breast Cancer | Olson D, Taylor J, Willis K, Hensley K, Allred S, Zaval M, Farr L, Thurman R, Jain N, Hein R, Ulrich M, Peterson S, Kulukian A | 2023 | Cancer Research Communications | Oncology | Classifier, Multiplex IHC | HALO |
| Tumor-associated macrophages trigger MAIT cell dysfunction at the HCC invasive margin | Ruf B, Bruhns M, Babaei S, Kedei N, Ma L, Revsine M, Benmebarek MR, Ma C, Heinrich B, Subramanyam V, Qi J, Wabitsch S, Green BL, Bauer KC, Myojin Y, Greten LT, McCallen JD, Huang P, Trehan R, Wang X, Nur A, Soika DQM, Pouzolles M, Evans CN, Chari R, Kleiner DE, Telford W, Dadkhah K, Ruchinskas A, Stovroff MK, Kang J, Oza K, Ruchirawat M, Kroemer A, Wang XW, Claassen M, Korangy F, Greten TF | 2023 | Cell | Immuno-oncology | Classifier, Spatial Analysis, Highplex FL, Figure Maker | HALO |
| Tumor cell HLA class I expression and pathologic response following neoadjuvant immunotherapy for newly diagnosed head and neck cancer | Robbins Y, Friedman J, Redman J, Sievers C, Lassoued W, Gulley J, Allen C | 2023 | Oral Oncology | Immuno-oncology | Classifier, Spatial Analysis, Highplex FL | HALO |
| Establishment of reference intervals of endometrial immune cells during the mid-luteal phase | Yu S, Huang C, Lian R, Diao L, Zhang X, Cai S, Wei H, Chen C, Li Y, Zeng Y | 2023 | Journal of Reproductive Immunology | Immunology | Classifier, Multiplex IHC | HALO |
| Anticoagulants Enhance Molecular and Cellular Immunotherapy of Cancer by Improving Tumor Microcirculation Structure and Function and Redistributing Tumor Infiltrates | Wei F, Su Y, Quan Y, Li X, Zou Q, Zhang L, Li S, Jiang M, Lin G, Liang P, He J, Xie K | 2023 | Clinical Cancer Research | Immuno-oncology | Classifier, Multiplex IHC, Spatial Analysis, Highplex FL | HALO |
| Apolipoprotein E induces pathogenic senescent-like myeloid cells in prostate cancer | Bancaro N, Cali B, Troiani M, Elia A, Arzola R, Attanasio G, Lai P, Crespo M, Gurel B, Pereira R, Guo C, Mosole S, Brina D, D'Ambrosio M, Pasquini E, Spataro C, Zagato E, Rinaldi A, Pedotti M, Lascio S, Meani F, Montopoli M, Ferrari M, Gallina A, Varani L, Mestre R, Bolis M, Sommer S, de Bono J, Calcinotto A, Alimonti A | 2023 | Cancer Cell | Oncology | Classifier, Highplex FL | HALO |
| Lineage tracing reveals clonal progenitors and long-term persistence of tumor-specific T cells during immune checkpoint blockade | Pai J, Hellmann M, Sauter J, Mattar M, Rizvi H, Woo H, Shah N, Nguyen E, Uddin F, Quintanal-Villalonga A, Chan J, Manoj P, Allaj V, Baine M, Bhanot U, Jain M, Linkov I, Meng F, Brown D, Chaft J, Plodkowski A, Gigoux M, Won H, Sen T, Wells D, Donoghue M, Stanchina E, Wolchok J, Loomis B, Merghoub T, Rudin C, Chow A, Satpathy A | 2023 | Cancer Cell | Immuno-oncology | Classifier | HALO |
| Immune profiling and RNA-seq uncover the cause of partial unexplained recurrent implantation failure | Fan X, Zhao Q, Li Y, Chen Z, Liao J, Chen H, Meng F, Lu G, Lin G, Gong F | 2023 | International Immunopharmacology | Immunology | Classifier, Multiplex IHC | HALO |
| Targeting the ATF6-mediated ER stress response and autophagy blocks integrin-driven prostate cancer progression | Macke A, Pachikov A, Divita T, Morris M, LaGrange C, Holzapfel M, Kubyshkin A, Zyablitskaya E, Makalish T, Eremenko S, Qiu H, Riethoven J, Hemstreet G, Petrosyan A | 2023 | Molecular Cancer Research | Oncology | Classifier, Area Quantification, Multiplex IHC, TMA | HALO |
| Comparing the peri-implantation endometrial T-bet/GATA3 ratio between control fertile women and patients with recurrent miscarriage: establishment and application of a reference range Get access Arrow | Yu S, Diao L, Lian R, Chen C, Huang C, Li X, Li Y, Zeng Y | 2023 | Human Reproduction | Other | Classifier, Multiplex IHC | HALO |
| Molecular determinants of clinical outcomes of pembrolizumab in recurrent ovarian cancer: Exploratory analysis of KEYNOTE-100 | Ledermann JA, Shapira-Frommer R, Santin AD, Lisyanskaya AS, Pignata S, Vergote I, Raspagliesi F, Sonke GS, Birrer M, Provencher DM, Sehouli J, Colombo N, González-Martín A, Oaknin A, Ottevanger PB, Rudaitis V, Kobie J, Nebozhyn M, Edmondson M, Sun Y, Cristescu R, Jelinic P, Keefe SM, Matulonis UA | 2023 | Gynecologic Oncology | Immuno-oncology | Classifier, Highplex FL | HALO |
| Classification of the tumor immune microenvironment and associations with outcomes in patients with metastatic melanoma treated with immunotherapies | Adegoke N, Gide T, Mao Y, Quek C, Patrick E, Carlino M, Lo S, Menzies A, da Silva P, Vergara I, Long G, Scolyer R, Wilmott J | 2023 | Journal for ImmunoTherapy of Cancer | Immuno-oncology | Classifier, Highplex FL | HALO |
| Empowering Renal Cancer Management with AI and Digital Pathology: Pathology, Diagnostics and Prognosis | Ivanova E, Fayzullin A, Grinin V, Ermilov D, Arutyunyan A, Timashev P, Shekhter A | 2023 | Biomedicines | Review | Classifier, Area Quantification, Object Colocalization | HALO, HALO AI |
| Deep radiomics-based fusion model for prediction of bevacizumab treatment response and outcome in patients with colorectal cancer liver metastases: a multicentre cohort study | Zhou S, Sun D, Mao W, Liu Y, Cen W, Ye L, Liang F, Xu J, Shi H, Ji Y, Wang L, Chang W | 2023 | eClinicalMedicine | Oncology | Classifier, Multiplex IHC | HALO |
| Unaltered hepatic wound healing response in male rats with ancestral liver injury | Beil J, Perner J, Pfaller L, Gérard M, Piaia A, Doelemeyer A, Wasserkrug Naor A, Martin L, Piequet A, Dubost V, Chibout S, Moggs J, Terranova R | 2023 | Nature Communications | Other | Classifier, Area Quantification | HALO, HALO AI |
| Disease pathology signatures in a mouse model of Mucopolysaccharidosis type IIIB | Petrova R, Patil AR, Trinh V, McElroy KE, Bhakta M, Tien J, Wilson DS, Warren L, Stratton JR | 2023 | Scientific Reports | Neuroscience, Metabolism | Classifier, Area Quantification, Object Colocalization | HALO |
| In vitro and In vivo evidence demonstrating chronic absence of Ref-1 Cysteine 65 impacts Ref-1 folding configuration, redox signaling, proliferation and metastasis in pancreatic cancer | Mijit M, Kpenu E, Chowdhury N, Gampala S, Wireman R, Liu S, Babb O, Georgiadis M, Wan J, Fishel M, Kelley M | 2023 | Redox Biology | Oncology | Classifier | HALO |
| The tumor-derived cytokine Chi3l1 induces neutrophil extracellular traps that promote T cell exclusion in triple-negative breast cancer | Taifour T, Attalla SS, Zuo D, Gu Y, Sanguin-Gendreau V, Proud H, Solymoss E, Bui T, Kuasne H, Papavasiliou V, Lee CG, Kamle S, Siegel PM, Elias JA, Park M, Muller WJ | 2023 | Immunity | Immuno-oncology | Classifier, Multiplex IHC, Spatial Analysis, Highplex FL | HALO |
| Components of the tumor immune microenvironment based on m-IHC correlate with prognosis and subtype of triple-negative breast cancer | Lin L, Li H, Wang X, Wang Z, Su G, Zhou J, Sun S, Ma X, Chen Y, You C, Gu Y | 2023 | Cancer Medicine | Immuno-oncology | Classifier, Spatial Analysis, Highplex FL | HALO |
| CD44 expression in renal tubular epithelial cells in the kidneys of rats with cyclosporine-induced chronic kidney disease | Matsushita K, Toyoda T, Akane H, Morikawa T, Ogawa K | 2023 | Journal of Toxicologic Pathology | Toxicology | Classifier | HALO |
| Integrated multi-omics profiling to dissect the spatiotemporal evolution of metastatic hepatocellular carcinoma | Sun Y, Wu P, Zhang Z, Wang Z, Zhou K, Song M, Ji Y, Zang F, Lou L, Rao K, Wang P, Gu Y, Gu J, Lu B, Chen L, Pan X, Zhao X, Peng L, Liu D, Chen X, Wu K, Lin P, Wu L, Su Y, Du M, Hou Y, Yang X, Qiu S, Shi Y, Sun H, Zhou J, Huang X, Peng D, Zhang L, Fan J | 2023 | Cancer Cell | Oncology | Classifier, Multiplex IHC, Spatial Analysis, Highplex FL | HALO |
| Tumor-associated fibrosis impairs immune surveillance and response to immune checkpoint blockade in non–small cell lung cancer | Herzog B, Baer J, Borcherding N, Kingston N, Belle J, Knolhoff B, Hogg G, Ahmad F, Kang L, Petrone J, Lin C, Govindan R, Denardo D | 2023 | Science Translational Medicine | Oncology | Classifier, Multiplex IHC | HALO |
| Analysis of several common APOBEC-type mutations in bladder tumors suggests links to viral infection | Rao N, Starrett G, Piaskowski M, Butler K, Golubeva Y, Yan W, Lawrence S, Dean M, Garcia-Closas M, Baris D, Johnson A, Schwenn M, Malats N, Real F, Kogevinas M, Rothman N, Silverman D, Dyrskjøt L, Buck C, Koutros S, Prokunina-Olsson L | 2023 | Cancer Prevention Research | Oncology | Classifier, Immune | HALO |
| Complex Urban Atmospheres alters Alveolar Stem Cells Niche Properties and drives Lung Fibrosis | Belgacemi R, Baptista BR, Justeau G, Toigo M, Frauenpreis A, Yilmaz R, Der Vartanian A, Cazaunau M, Pangui E, Bergé A, Gratien A, Macias Rodriguez JC, Bellusci S, Derumeaux G, Boczkowski J, Al Alam D, Coll P, Lanone S, Boyer L | 2023 | American Journal of Physiology - Lung Cellular and Molecular Physiology | Fibrosis | Classifier, Area Quantification, Highplex FL | HALO |
| The tumor microenvironment state associates with response to HPV therapeutic vaccination in patients with respiratory papillomatosis | NORBERG S, BAI K, SIEVERS C, ROBBINS Y, FRIEDMAN J, YANG X, KENYON M, WARD E, SCHLOM J, GULLEY J, LANKFORD A, SEMNANI R, SABZEVARI H, BROUGH D, ALLEN C | 2023 | Science Translational Medicine | Oncology | Classifier, Highplex FL | HALO, HALO AI |
| Role of CD44 expressed in renal tubules during maladaptive repair in renal fibrogenesis in an allopurinol-induced rat model of chronic kidney disease | Matsushita K, Toyoda T, Akane H, Morikawa T, Ogawa K | 2023 | Journal of Applied Toxicology | Toxicology | Classifier | HALO |
| Exocrine pancreas in type 1 and type 2 diabetes: different patterns of fibrosis, metaplasia, angiopathy, and adiposity | Wright J, Eskaros A, Windon A, Bottino R, Jenkins R, Bradley A, Aramandla R, Philips S, Kang H, Saunders D, Brissova M, Powers A | 2023 | Diabetes | Gastroenterology | Classifier, Highplex FL | HALO |
| A modified mouse model of Friedreich's ataxia with conditional Fxn allele homozygosity delays onset of cardiomyopathy | Perfitt TL, Huichalaf C, Gooch R, Kuperman A, Ahn Y, Chen X, Ullas S, Hirenallur-Shanthappa D, Zhan Y, Otis D, Whiteley LO, Bulawa C, Martelli A | 2023 | American Journal of Physiology-Heart and Circulatory Physiology | Other | Classifier, Area Quantification | HALO |
| Mutant p53 gains oncogenic functions through a chromosomal instability-induced cytosolic DNA response | Zhao M, Wang T, Gleber-Netto F O, Chen Z, McGrail D J, Gomez J A, Ju W, Gadhikar M A, Ma W, Shen L, Wang Q, Tang X, Pathak S, Raso M G, Burks J K, Lin S-Y, Wang J, Multani A S, Pickering C R, Chen J, Myers J N, Zhou G | 2024 | Nature Communications | Oncology | Classifier, Multiplex IHC | HALO |
| Early immune remodeling steers clinical response to frontline chemoimmunotherapy in advanced gastric cancer | An M, Mehta A, Min BH, Geo YJ, Wright SJ, Parikh M, Bi L, Lee H, Kim TJ, Lee SY, Moon J, Park RJ, Strickland MR, Park WY, Kang WK, Kim KM, Kim ST, Lempner SJ, Lee J | 2024 | Cancer Discovery | Immuno-oncology | Classifier, Highplex FL | HALO |
| Nanoparticle mediated photodynamic therapy as a method to ablate oral cavity squamous cell carcinoma in preclinical models | Sahovaler A, Valic M, Townson J, Chan H, Zheng M, Tzelnick S, Mondello T, Pener-Tessler A, Eu D, El-Sayes A, Ding L, Chen J, Douglas C, Weersink R, Muhanna N, Zheng G, Irish J | 2024 | Cancer Research | Oncology | Classifier, Multiplex IHC | HALO |
| Translational and Therapeutic Evaluation of RAS-GTP Inhibition by RMC-6236 in RAS-Driven Cancers | Jiang J, Jiang L, Maldonato B J, Wang Y, Holderfield M, Aronchik I, Winters I P, Salman Z, Blaj C, Menard M, Brodbeck J, Chen Z, Wei X, Rosen M J, Gindin Y, Lee B J, Evans J W, Chang S, Wang Z, Seamon K J, Parsons D, Cregg J, Marquez A, Tomlinson A C A, Yano J K, Knox J E, Quintana E, Aguirre A J, Arbour K C, Reed A, Gustafson W C, Gill A L, Koltun E S, Wildes D, Smith J A M, Wang Z, Singh M | 2024 | Cancer Discovery | Oncology | Classifier, Area Quantification, Cytonuclear | HALO |
| Tertiary lymphoid structures and B cells determine clinically relevant T cell phenotypes in ovarian cancer | Kasikova L, Rakova J, Hensler M, Lanickova T, Tomankova J, Pasulka J, Drozenova J, Mojzisova K, Fialova A, Vosahlikova S, Laco J, Ryska A, Dundr P, Kocian R, Brtnicky T, Skapa P, Capkova L, Kovar M, Prochazka J, Praznovec I, Koblizek V, Taskova A, Tanaka H, Lischke R, Mendez F C, Jr J V, Heinzelmann-Schwarz V, Jacob F, McNeish I A, Halaska M J, Rob L, Cibula D, Orsulic S, Galluzzi L, Spisek R, Fucikova J | 2024 | Nature Communications | Oncology | Classifier, Highplex FL, Registration | HALO |
| Obesogenic High-Fat Diet and MYC Cooperate to Promote Lactate Accumulation and Tumor Microenvironment Remodeling in Prostate Cancer | Boufaied N, Chetta P, Hallal T, Cacciatore S, Lalli D, Luthold C, Homsy K, Imada E, Syamala S, Photopoulos C, Di Matteo A, de Polo A, Storaci A, Huang Y, Giunchi F, Sheridan P, Michelotti G, Nguyen Q, Zhao X, Liu Y, Davicioni E, Spratt D, Sabbioneda S, Maga G, Mucci L, Ghigna C, Marchionni L, Butler L, Ellis L, Bordeleau F, Loda M, Vaira V, Labbé D, Zadra G | 2024 | Cancer Research | Oncology | Classifier, Multiplex IHC | HALO |
| Adjuvant COX inhibition augments STING signaling and cytolytic T cell infiltration in irradiated 4T1 tumors | Ridnour L, Cheng R, Kedei N, Somasundaram V, Bhattacharyya D, Basudhar D, Wink A, Walke A, Kim C, Heinz W, Edmondson E, Butcher D, Warner A, Dorsey T, Pore M, Kinders R, Lipkowitz S, Bryant R, Rittscher J, Wong S, Hewitt S, Chang J, Shalaby A, Callagy G, Glynn S, Ambs S, Anderson S, McVicar D, Lockett S, Wink D | 2024 | JCI Insight | Immuno-oncology | Classifier, FISH, Spatial Analysis, Highplex FL | HALO |
| Development of human pancreatic cancer avatars as a model for dynamic immune landscape profiling and personalized therapy | Hughes D, Evans A, Go S, Eyres M, Pan L, Mukherjee S, Soonawalla Z, Willenbrock F, O'Neill E | 2024 | Science Advances | Immuno-oncology | Classifier, Spatial Analysis, Highplex FL | HALO |
| Combined inhibition of KRASG12C and mTORC1 kinase is synergistic in non-small cell lung cancer | Kitai H, Choi P H, Yang Y C, Boyer J A, Whaley A, Pancholi P, Thant C, Reiter J, Chen K, Markov V, Taniguchi H, Yamaguchi R, Ebi H, Evans J, Jiang J J, Lee B, Wildes D, Stanchina E, Smith J A M, Singh M, Rosen N | 2024 | Nature Communications | Oncology | Classifier, Area Quantification, Mulitplex iHC | HALO |
| PD‐L1+ macrophage and tumor cell abundance and proximity to T cells in the pretreatment large B‐cell lymphoma microenvironment impact CD19 CAR‐T cell immunotherapy efficacy | Hirayama A V, Wright J H, Smythe K S, Fiorenza S, Shaw A N, Gauthier J, Maloney D G, Naresh K N, Yeung C C S, Turtle C J | 2024 | Hemasphere | Immuno-oncology | Classifier, Spatial Analysis, Highplex FL | HALO, HALO Link |
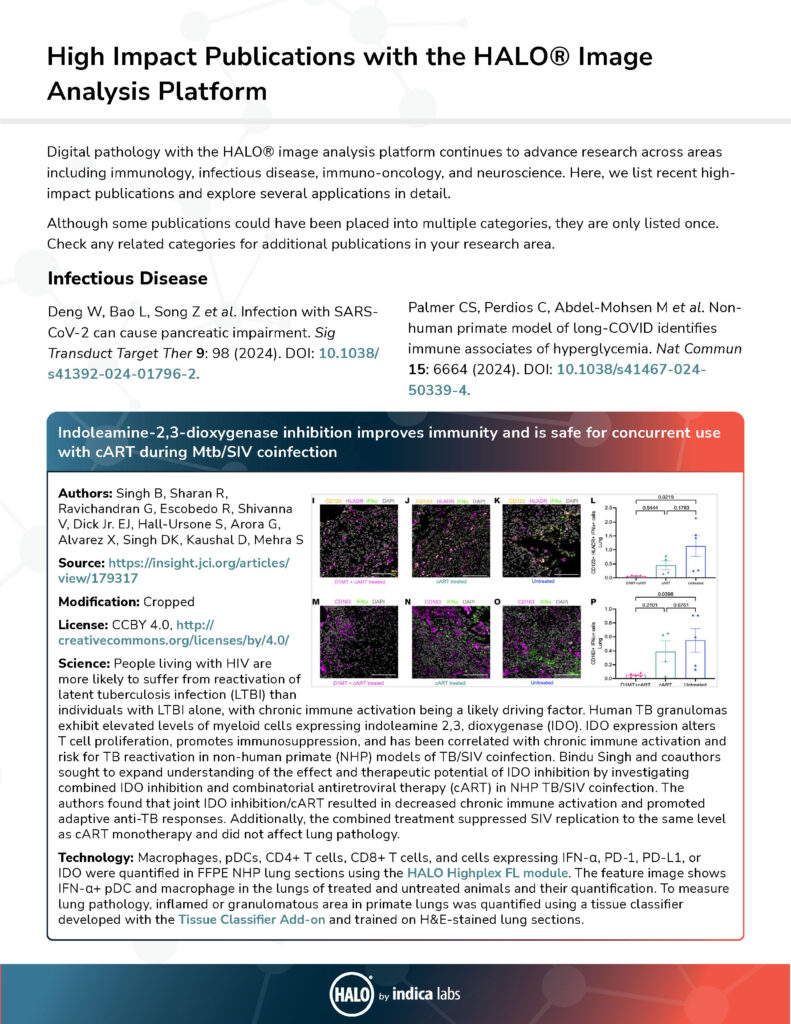
| Indoleamine-2,3-dioxygenase inhibition improves immunity and is safe for concurrent use with cART during Mtb/SIV coinfection | Singh B, Sharan R, Ravichandran G, Escobedo R, Shivanna V, Dick E J, Hall-Ursone S, Arora G, Alvarez X, Singh D K, Kaushal D, Mehra S | 2024 | JCI Insight | Infectious Disease | Classifier, Highplex FL | HALO |
| Chemotherapy drives tertiary lymphoid structures that correlate with ICI-responsive TCF1+CD8+ T cells in metastatic ovarian cancer | Lanickova T, Hensler M, Kasikova L, Vosahlikova S, Angelidou A, Pasulka J, Griebler H, Drozenova J, Mojzisova K, Vankerckhoven A, Laco J, Ryška A, Dundr P, Kocian R, Cibula D, Brtnicky T, Skapa P, Jacob F, Kovar M, Praznovec I, McNeish I A, Halaska M J, Rob L, Coosemans A, Orsulic S, Galluzzi L, Spisek R, Fucikova J | 2024 | Clinical Cancer Research | Immuno-oncology | Classifier, Highplex FL, Registration | HALO |
| Diversity of the immune microenvironment and response to checkpoint inhibitor immunotherapy in mucosal melanoma | Vos J L, Traets J J H, Qiao X, Seignette I M, Peters D, Wouters M W J M, Hooijberg E, Broeks A, van der Wal J E, Karakullukcu M B, Klop W M C, Navran A, van Beurden M, Brouwer O R, Morris L G T, van Poelgeest M I E, Kapiteijn E, Haanen J B A G, Blank C U, Zuur C L | 2024 | JCI Insight | Immuno-oncology | Classifier, Spatial Analysis, Highplex FL | HALO |
| https://www.cell.com/cell-reports-medicine/fulltext/S2666-3791(24)00623-2?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS2666379124006232%3Fshowall%3Dtrue | Chakraborty S, Sen U, Ventura K, Jethalia V, Coleman C, Sridhar S, Banerjee A, Ozakinci H, Mahendravarman Y, Snioch K, Stanchina E, Shields M D, Tomalin L E, Demircioglu D, Boyle T A, Tocheva A, Hasson D, Sen T | 2024 | Cell Reports Medicine | Immuno-oncology | Classifier, Highplex FL | HALO |
| Hypoxia promotes tumor immune evasion by suppressing MHC-I expression and antigen presentation | Estephan H, Tailor A, Parker R, Kreamer M, Papandreou I, Campo L, Easton A, Moon EJ, Denko NC, Ternette N, Hammond EM, Giaccia AJ | 2025 | The EMBO Journal | Immuno-oncology | Classifier, Area Quantification, Spatial Analysis, Highplex FL | HALO |
| Efficacy and tolerability of neoadjuvant therapy with Talimogene laherparepvec in cutaneous basal cell carcinoma: a phase II trial (NeoBCC trial) | Ressler J M, Plaschka M, Silmbrod R, Bachmayr V, Shaw L E, Silly T, Zila N, Stepan A, Kusienicka A, Tschandl P, Tittes J, Roka F, Haslik W, Petzelbauer P, Koenig F, Kunstfeld R, Farlik M, Halbritter F, Weninger W, Hoeller C | 2025 | Nature Cancer | Oncology | Classifier, Spatial Analysis, Highplex FL | HALO, HALO AI |
| A multistage drug delivery approach for colorectal primary tumors and lymph node metastases | Yuan Y, Lin Q, Feng HY, Zhang Y, Lai X, Zhu MH, Wang J, Shi J, Huang Y, Zhang L, Lu Q, Yuan Z, Lovell JF, Chen HZ, Sun P, Fang C | 2025 | Nature Communications | Oncology | Classifier, Area Quantification, Spatial Analysis | HALO |
| Endothelial cell pY397-FAK expression predicts risk of breast cancer recurrences after radiotherapy in SweBCG91-RT cohort | Drake R J, Landén A H, Holmberg E, Stenmark Tullberg A, Killander F, Niméus E, Jordan A, McGuinness J, Karlsson P, Hodivala-Dilke K | 2025 | Clinical Cancer Research | Oncology | Classifier, Area Quantification | HALO |
| Distinct immune cell infiltration patterns in pancreatic ductal adenocarcinoma (PDAC) exhibit divergent immune cell selection and immunosuppressive mechanisms | Sivakumar S, Jainarayanan A, Arbe-Barnes E, Sharma P K, Leathlobhair M N, Amin S, Reiss D J, Heij L, Hegde S, Magen A, Tucci F, Sun B, Wu S, Anand N M, Slawinski H, Revale S, Nassiri I, Webber J, Hoeltzel G D, Frampton A E, Wiltberger G, Neumann U, Charlton P, Spiers L, Elliott T, Wang M, Couto S, Lila T, Sivakumar P V, Ratushny A V, Middleton M R, Peppa D, Fairfax B, Merad M, Dustin M L, Abu-Shah E, Bashford-Rogers R | 2025 | Nature Communications | Immuno-oncology | Classifier, Multiplex IHC | HALO |
| Dietary Oxidized Lipids in Redox Biology: Oxidized Olive Oil Disrupts Lipid Metabolism and Induces Intestinal and Hepatic Inflammation in C57BL/6J Mice | Bao Y, Osowiecka M, Ott C, Tziraki V, Meusburger L, Blaßnig C, Krivda D, Pjevac P, Silva J, Strauss M, Steffen C, Heck V, Aygün S, Duszka K, Doppelmayer K, Grune T, Pignitter M | 2025 | Redox Biology | Metabolism | Classifier, Area Quantification | HALO |
| CD4+ T helper 2 cell–macrophage crosstalk induces IL-24–mediated breast cancer suppression | Wang B, Xia Y, Zhou C, Zeng Y, Son HG, Demehri S | 2025 | JCI Insight | Immuno-oncology | Highplex FL | HALO, HALO AI |
| Efficacy and tolerability of neoadjuvant therapy with Talimogene laherparepvec in cutaneous basal cell carcinoma: a phase II trial (NeoBCC trial) | Ressler J M, Plaschka M, Silmbrod R, Bachmayr V, Shaw L E, Silly T, Zila N, Stepan A, Kusienicka A, Tschandl P, Tittes J, Roka F, Haslik W, Petzelbauer P, Koenig F, Kunstfeld R, Farlik M, Halbritter F, Weninger W, Hoeller C | 2025 | Nature Cancer | Oncology | Classifier, Spatial Analysis, Highplex FL | HALO, HALO AI |
| A multistage drug delivery approach for colorectal primary tumors and lymph node metastases | Yuan Y, Lin Q, Feng HY, Zhang Y, Lai X, Zhu MH, Wang J, Shi J, Huang Y, Zhang L, Lu Q, Yuan Z, Lovell JF, Chen HZ, Sun P, Fang C | 2025 | Nature Communications | Oncology | Classifier, Area Quantification, Spatial Analysis | HALO |
| Endothelial cell pY397-FAK expression predicts risk of breast cancer recurrences after radiotherapy in SweBCG91-RT cohort | Drake R J, Landén A H, Holmberg E, Stenmark Tullberg A, Killander F, Niméus E, Jordan A, McGuinness J, Karlsson P, Hodivala-Dilke K | 2025 | Clinical Cancer Research | Oncology | Classifier, Area Quantification | HALO |
| Distinct immune cell infiltration patterns in pancreatic ductal adenocarcinoma (PDAC) exhibit divergent immune cell selection and immunosuppressive mechanisms | Sivakumar S, Jainarayanan A, Arbe-Barnes E, Sharma P K, Leathlobhair M N, Amin S, Reiss D J, Heij L, Hegde S, Magen A, Tucci F, Sun B, Wu S, Anand N M, Slawinski H, Revale S, Nassiri I, Webber J, Hoeltzel G D, Frampton A E, Wiltberger G, Neumann U, Charlton P, Spiers L, Elliott T, Wang M, Couto S, Lila T, Sivakumar P V, Ratushny A V, Middleton M R, Peppa D, Fairfax B, Merad M, Dustin M L, Abu-Shah E, Bashford-Rogers R | 2025 | Nature Communications | Immuno-oncology | Classifier, Multiplex IHC | HALO |
| Dietary Oxidized Lipids in Redox Biology: Oxidized Olive Oil Disrupts Lipid Metabolism and Induces Intestinal and Hepatic Inflammation in C57BL/6J Mice | Bao Y, Osowiecka M, Ott C, Tziraki V, Meusburger L, Blaßnig C, Krivda D, Pjevac P, Silva J, Strauss M, Steffen C, Heck V, Aygün S, Duszka K, Doppelmayer K, Grune T, Pignitter M | 2025 | Redox Biology | Metabolism | Classifier, Area Quantification | HALO |
Related HALO Modules
Quantify expression of an unlimited number of biomarkers in any cellular compartment - membrane, nucleus or cytoplasm.
Learn MoreQuantify expression of up to five brightfield stains in any cellular compartment - membrane, nucleus or cytoplasm.
Learn MorePlot cells and objects from one or more images and perform nearest neighbor analysis, proximity analysis, and tumor infiltration analysis.
Learn MoreUse the arrows above to view additional related modules
Want to Learn More?
Fill out the form below to request information about any of our software products.
You can also drop us an email at info@indicalab.com
Products & Services
Interested in purchasing or learning more about our products and services? Our highly trained application scientists are a couple of clicks away.
Software Maintenance & Support Coverage
Interested in purchasing an SMS plan? We would be happy to give you a quote.
Technical Support
Need technical support? Our IT specialists are here to help.






