Advancing Neuroscience Research with HALO® and HALO AI

- By Ben Dyer
- December 4th, 2025
As scientists work to continue unraveling the structural, cellular, and molecular complexities of the brain and its diseases, the characteristics of this unique tissue pose significant challenges to traditional histological analysis. Further complicating the issue, neuroscience studies increasingly leverage large datasets, multiplexed imaging, and multimodal quantification, producing large volumes of data that demand computational analysis.
Within this tissue and workflow context, digital pathology has become essential, with advanced platforms such as HALO® and HALO AI helping uncover biologically meaningful insights. Since 2015, HALO platforms have been cited in over 200 peer-reviewed neuroscience publications, helping to advance studies covering the range of topics in the field. In this blog post, we explore the hurdles associated with brain tissue analysis, discuss how HALO and HALO AI address these challenges, and highlight specific neuroscience applications where these tools are helping to drive discovery.
Challenges of Brain Tissue Image Analysis
From high-density cortical regions to heterogeneous inflammatory lesions, brain tissue is among the most complex biological substrates to analyze, with multiple characteristics that make quantitative image analysis particularly challenging. Brain sections contain a rich mixture of interspersed neuronal, glial, vascular, and immune cells, each differing in size, staining pattern, and frequency. These mixed populations often exist within regions of extremely high cellular density, such as the hippocampus or laminated cortical layers, where overlapping nuclei and processes make manual evaluation slow and even traditional quantitative image analysis error prone.
Many of these neural cell types exhibit highly irregular shapes, such as the branched axons and dendrites of neurons and the diverse morphologies of microglia. Adding further complexity, brain tissue itself exhibits intricate and varied morphology, from curved cortical laminae to region-specific cytoarchitecture, and disease-associated pathologies can introduce even more structural variability. Together, this cellular and tissue variety necessitates accurate, detailed segmentation that has historically required time-intensive assessment and annotation.
Benefits of Quantitative Digital Pathology in Neuroscience
Our HALO image analysis platform and HALO AI deep learning toolset are built for AI-powered quantitative analysis of whole slide images. These solutions provide a range of features for brightfield and fluorescence image analysis that empower researchers to meet the challenges of today’s neuroscience studies.
HALO and HALO AI offer pre-trained and custom AI models, respectively, for optimized nuclear and membrane segmentation. These AI-powered solutions enable accurate delineation of the densely packed, irregularly shaped, and diversely stained cells common in brain tissue sections. Pre-trained models provide significant improvement over traditional segmentation approaches, while models trained within HALO AI enable optimization for users’ specific applications.
Additionally, HALO AI allows researchers to develop models for automated tissue classification and object classification and phenotyping. Such models can streamline and standardize analysis of complex brain tissues, enabling automated quantification of different tissues, cells, or other objects of interest. Object phenotypers are especially well suited for neuroscience research, enabling automated classification of cells based on morphology, intra-cellular staining patterns, and other fine details that characterize the diverse cell populations of the nervous system.
Though AI models can run in isolation within HALO, they are frequently incorporated into HALO modules to further expand analysis options and readouts. HALO modules range from general-use solutions, like Highplex FL and ISH-IHC, to more specific modules, such as Microglial Activation and Axon Quantification. Depending on the module, readouts can include per-cell biomarker expression levels, object size and colocalization metrics, spatial distribution measurements, and more. In addition, modules, and individual AI models, can be run on images in batch, providing users with extensive, objective data to power their neuroscience studies.
HALO also offers a range of modules and add-ons that streamline workflows and further expand analysis capabilities. Add-ons like Tissue Microarray and Serial Section Analysis enable time- and tissue-saving workflows and fully integrate with HALO modules for analysis. The High Dimensional Analysis module offers dimensionality reduction and unsupervised clustering tools with interactive plotting natively within HALO, helping researchers explore and understand complex datasets faster.
Together, these features provide researchers with an accurate, reproducible, and highly scalable image analysis platform for extracting insights from even the most complex brain tissue datasets. In the following examples, we explore how HALO and HALO AI capabilities are being applied to answer diverse questions in neuroscience.
Nuclear Segmentation
As in all tissues, reliable nuclear segmentation is foundational for quantifying protein or RNA biomarkers, evaluating cellular phenotypes, and comparing disease states in the brain. Published examples of AI-powered nuclear segmentation abound, with Greally et al. leveraging a custom nuclear segmentation model for cells in the dorsolateral prefrontal cortex.1 Beyond their usual application, nuclear segmentation models can also be used for alternative applications, such as detecting biomarker-positive cells. For example, Butler et al. used a nuclear segmentation model to detect and quantify Nissl-positive neurons in the brain parenchyma while investigating neuron loss associated with repetitive head impacts.2
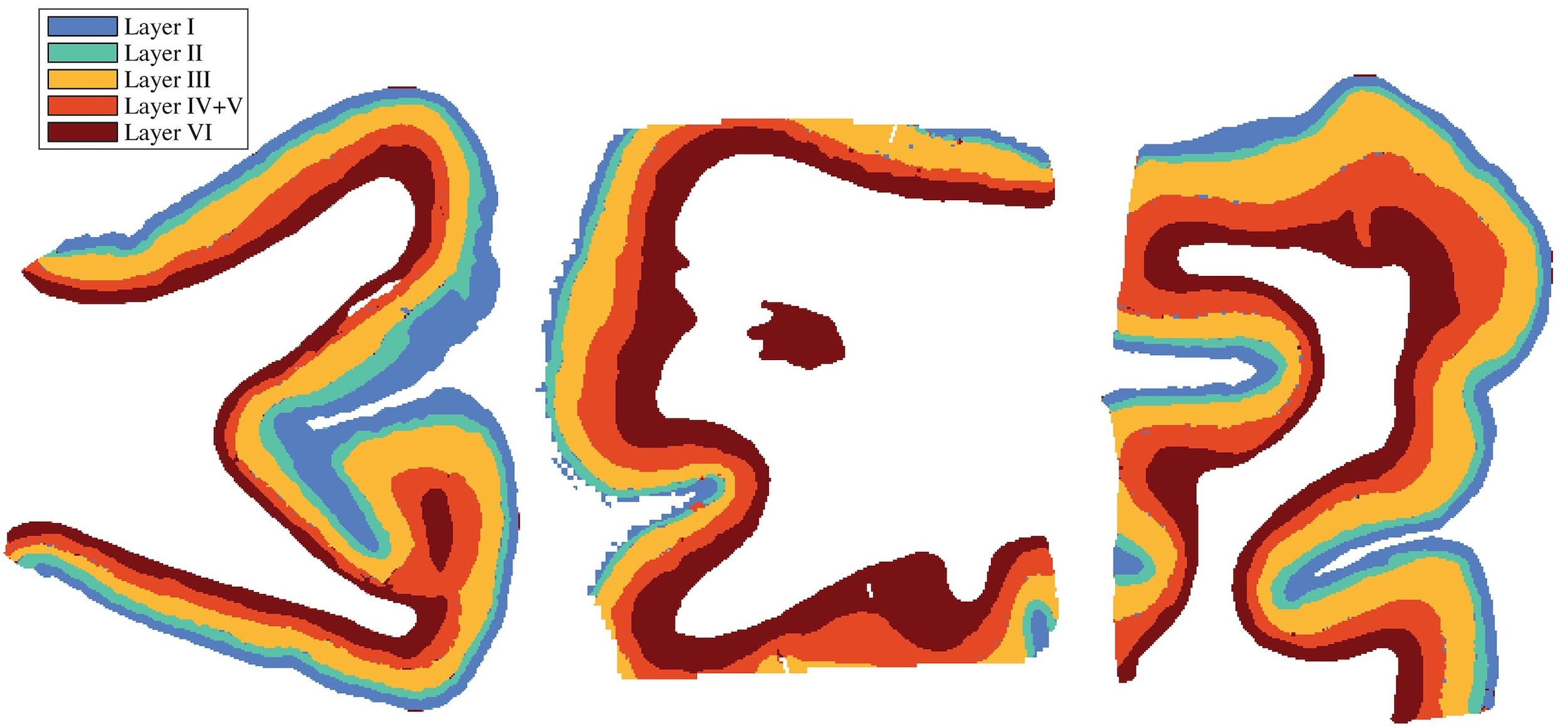
Tissue Segmentation
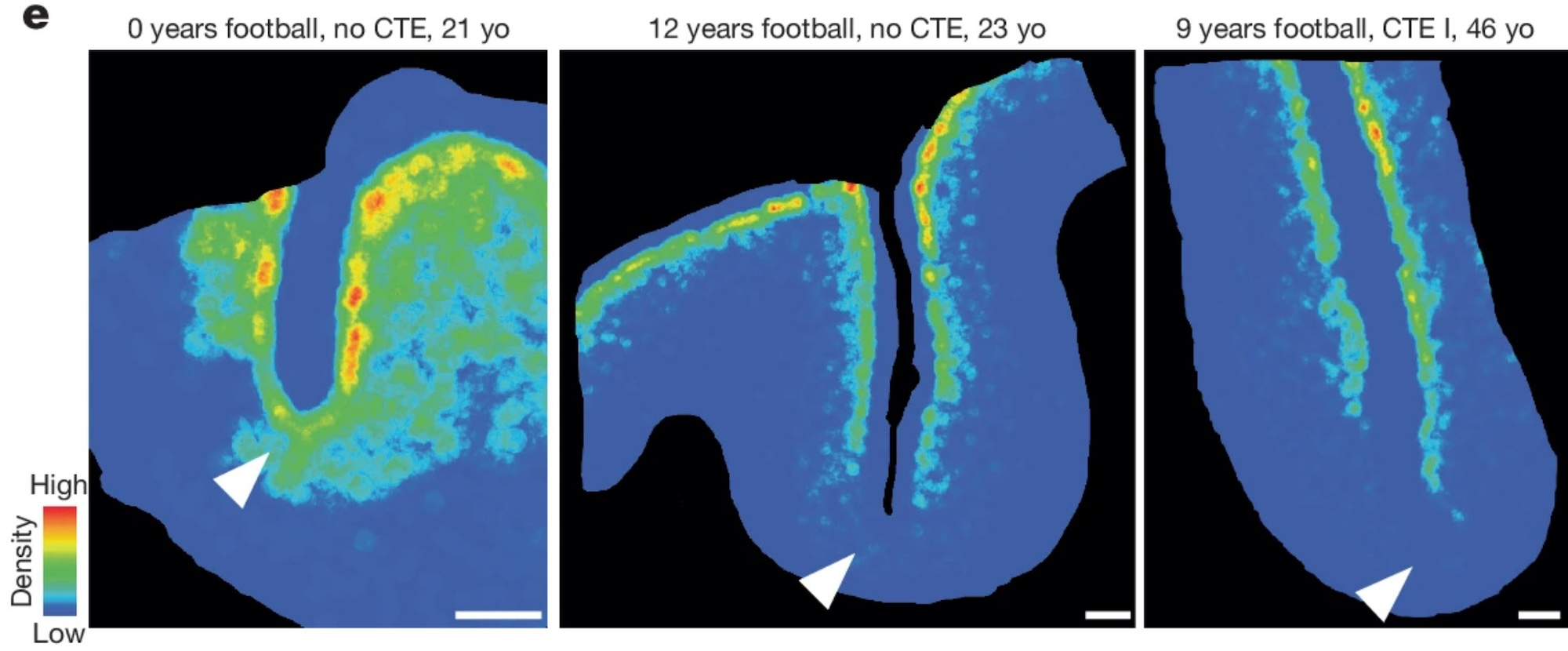
AI-powered tissue segmentation opens opportunities for structural mapping, regional analysis, and automated quantification across complex brain regions. Using HALO AI, Kundu et al. developed a tissue classifier for cortical layers I through VI, with the output used as a comparison for their novel MRI-based imaging modality.3 In their study of chronic traumatic encephalopathy endotypes, Han et al. used a custom HALO AI tissue classifier to quantify cortical gray matter area and standardize biomarker expression per area.4
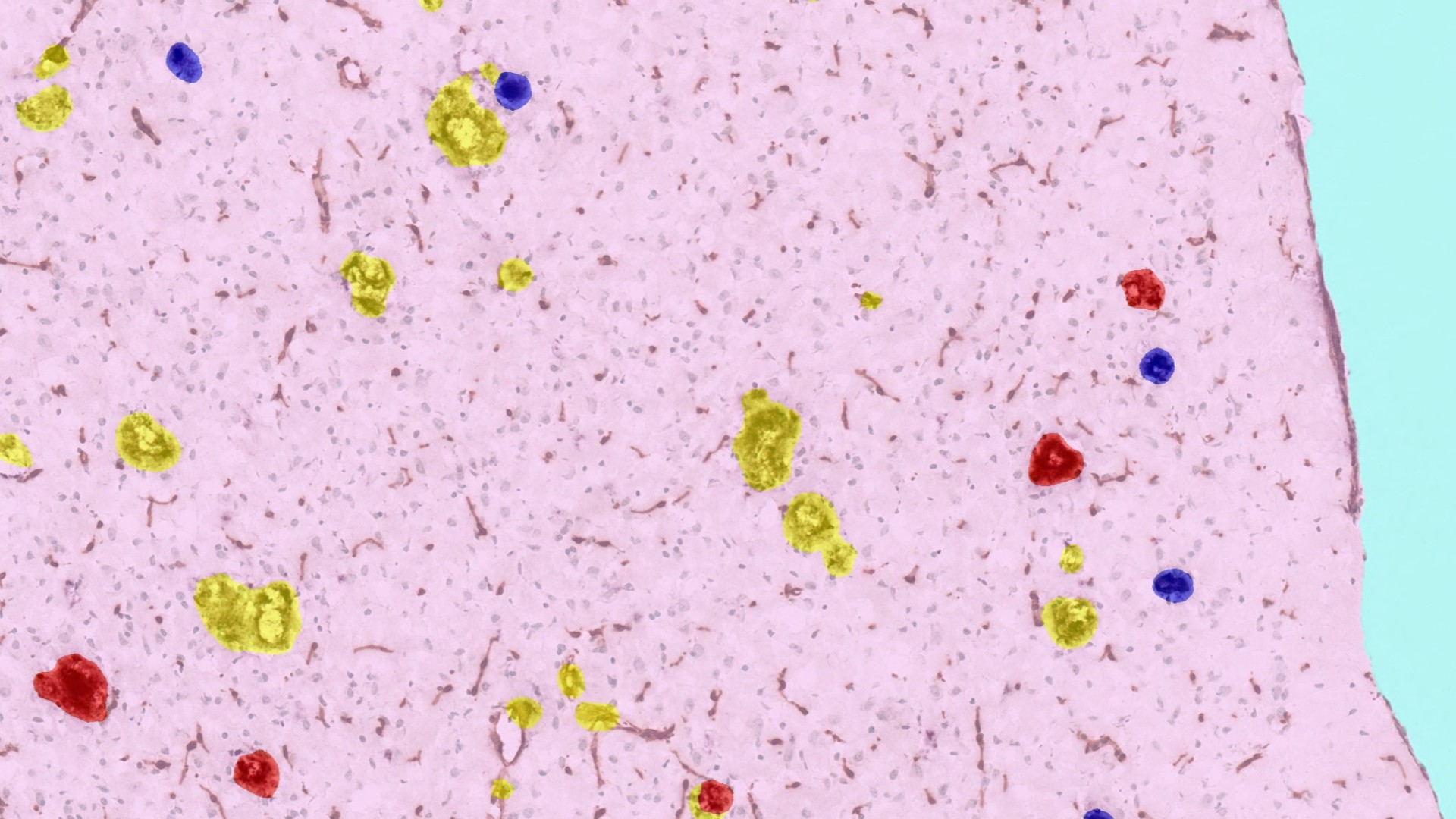
AI-Based Object Detection and Phenotyping
AI-based object detection and phenotyping enable identification and quantification of diverse structures or cells, streamlining a process that can otherwise be tedious and error prone. During their study of potential therapies for Alzheimer’s disease, Schauer et al. developed a classifier to quantify IBA1-stained microglia clusters across brain hemisphere sections.5 In the previously mentioned study, Han et al. leveraged a custom object phenotyper to automatically identify and quantify tau-positive cells in the gray matter identified by their tissue classifier.4
Biomarker Quantification
For both brightfield or fluorescence images, HALO modules offer a complete toolkit for analyzing biomarkers in either cell- or object-based analysis of brain tissue. In their study of repeated head trauma, Butler et al. leveraged multiple HALO modules to measure cellular RNA and protein expression across cells in the frontal cortex.2 In an object-based analysis of biomarkers, Shahidehpour et al. used the HALO Object Colocalization module to characterize TDP-43+ structures in middle frontal gyrus cortical gray matter as part of their study of limbic-predominant age-related TDP-43 encephalopathy neuropathologic change.6
Spatial Analysis
Spatial context is essential for understanding neural circuits, neuroinflammation, and progression of neurodegenerative disease. The HALO Spatial Analysis module provides a suite of tools for identifying proximity and relative spatial distribution of cells and objects. Using this module, Butler et al. quantified microglia/vessel pairs within 25 µm and analyzed neuron density in frontal cortex sulci.2
Conclusions
As neuroscience research continues to push the boundaries of what can be measured and understood within the brain, the demands upon quantitative image analysis have never been greater. Offering a flexible AI toolset, extensive analytics, and reliable results, HALO and HALO AI meet these challenges and are helping to accelerate discovery across neurodegeneration, traumatic brain injury, and beyond.
No matter what your neuroscience research area, HALO and HALO AI provide the precision, scalability, and analytical depth needed to advance your image analysis. If you’d like to learn more or discuss a specific research application, contact us at Info@IndicaLab.com. Additionally, visit our website to browse the features of the HALO modules most frequently used in neuroscience research, including the ISH, FISH, ISH-IHC, FISH-IF, Area Quantification, and Object Colocalization modules.
References
- Greally S, Kumar M, Schlaffner C et al. Dementia with lewy bodies patients with high tau levels display unique proteome profiles. Mol Neurodegener 19, 98 (2024). DOI: 1186/s13024-024-00782-0.
- Butler MLMD, Pervaiz N, Breen K et al. Repeated head trauma causes neuron loss and inflammation in young athletes. Nature 647, 228-237 (2025). DOI: 1038/s41586-025-09534-6.
- Kundu S, Barsoum S, Ariza J et al. Mapping the individual human cortex using multidimensional MRI and unsupervised learning. Brain Commun 5(6) (2023). DOI: 1093/braincomms/fcad258.
- Han X, Zhang Y, Petrosky JN et al. A structural haplotype in the 17q21.31 MAPT region is associated with increased risk for chronic traumatic encephalopathy endophenotypes. Cell Rep Med 6(5) (2025). DOI: 1016/j.xcrm.2025.102084.
- Schauer SP, Cho CH, Novikova G et al. Primate cerebrospinal fluid CHI3L1 reflects brain TREM2 agonism. Alzheimers Dement 20(9), 5861-5888 (2024). DOI: 1002%2Falz.13921.
- Shahidehpour RK, Katsumata Y, Dickson DW et al. LATE-NC Stage 3: a diagnostic rubric to differentiate severe LATE-NC from FTLD-TDP. Acta Neuropathol 149, 38 (2025). DOI: 1007/s00401-025-02876-5.